Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
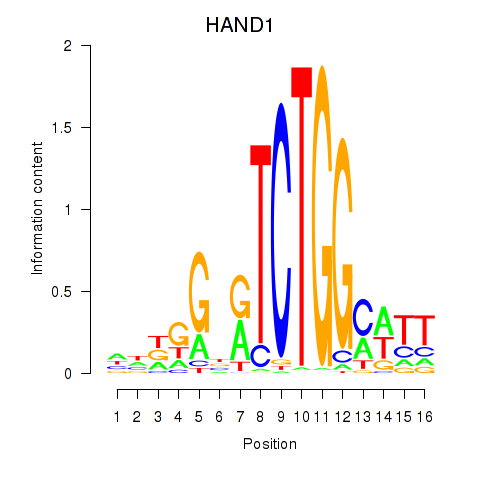
Results for HAND1
Z-value: 0.22
Transcription factors associated with HAND1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HAND1
|
ENSG00000113196.3 | heart and neural crest derivatives expressed 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HAND1 | hg38_v1_chr5_-_154478218_154478234 | 0.40 | 3.3e-01 | Click! |
Activity profile of HAND1 motif
Sorted Z-values of HAND1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_148697784 | 0.22 |
ENST00000497524.5
ENST00000418473.7 ENST00000349243.8 ENST00000404754.2 |
AGTR1
|
angiotensin II receptor type 1 |
| chr19_+_12751789 | 0.22 |
ENST00000553030.6
|
BEST2
|
bestrophin 2 |
| chr6_+_101181254 | 0.21 |
ENST00000682090.1
ENST00000421544.6 |
GRIK2
|
glutamate ionotropic receptor kainate type subunit 2 |
| chr19_+_12751672 | 0.20 |
ENST00000549706.5
|
BEST2
|
bestrophin 2 |
| chr2_+_209579399 | 0.20 |
ENST00000360351.8
|
MAP2
|
microtubule associated protein 2 |
| chr2_+_209579598 | 0.20 |
ENST00000445941.5
ENST00000673860.1 |
MAP2
|
microtubule associated protein 2 |
| chr6_+_123803853 | 0.17 |
ENST00000368417.6
|
NKAIN2
|
sodium/potassium transporting ATPase interacting 2 |
| chr2_+_209579429 | 0.13 |
ENST00000361559.8
|
MAP2
|
microtubule associated protein 2 |
| chr3_+_141384790 | 0.13 |
ENST00000507722.5
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr20_-_1184981 | 0.12 |
ENST00000429036.2
|
TMEM74B
|
transmembrane protein 74B |
| chr19_+_35868585 | 0.11 |
ENST00000652533.1
|
APLP1
|
amyloid beta precursor like protein 1 |
| chr19_+_35868518 | 0.11 |
ENST00000221891.9
|
APLP1
|
amyloid beta precursor like protein 1 |
| chrX_+_150363306 | 0.10 |
ENST00000370401.7
ENST00000432680.7 |
MAMLD1
|
mastermind like domain containing 1 |
| chrX_+_150363258 | 0.10 |
ENST00000683696.1
|
MAMLD1
|
mastermind like domain containing 1 |
| chr5_-_147906530 | 0.10 |
ENST00000318315.5
ENST00000515291.1 |
C5orf46
|
chromosome 5 open reading frame 46 |
| chr8_+_78516329 | 0.09 |
ENST00000396418.7
ENST00000352966.9 |
PKIA
|
cAMP-dependent protein kinase inhibitor alpha |
| chr1_+_65309517 | 0.09 |
ENST00000371069.5
|
DNAJC6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr8_-_92017637 | 0.08 |
ENST00000422361.6
|
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1 |
| chr3_+_51861604 | 0.08 |
ENST00000333127.4
|
IQCF2
|
IQ motif containing F2 |
| chr4_+_41613476 | 0.08 |
ENST00000508466.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr7_-_81770122 | 0.08 |
ENST00000423064.7
|
HGF
|
hepatocyte growth factor |
| chr6_-_39314445 | 0.08 |
ENST00000453413.2
|
KCNK17
|
potassium two pore domain channel subfamily K member 17 |
| chr6_-_39314412 | 0.08 |
ENST00000373231.9
|
KCNK17
|
potassium two pore domain channel subfamily K member 17 |
| chr4_-_25030922 | 0.08 |
ENST00000382114.9
|
LGI2
|
leucine rich repeat LGI family member 2 |
| chr1_-_110064916 | 0.08 |
ENST00000649954.1
|
ALX3
|
ALX homeobox 3 |
| chr8_+_30744150 | 0.07 |
ENST00000265616.10
ENST00000341403.9 ENST00000615729.4 |
UBXN8
|
UBX domain protein 8 |
| chr4_+_41612702 | 0.07 |
ENST00000509277.5
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr11_+_101914997 | 0.07 |
ENST00000263468.13
|
CEP126
|
centrosomal protein 126 |
| chrX_-_34657274 | 0.07 |
ENST00000275954.4
|
TMEM47
|
transmembrane protein 47 |
| chr20_+_31368594 | 0.07 |
ENST00000253381.3
|
DEFB118
|
defensin beta 118 |
| chr2_+_201132958 | 0.06 |
ENST00000479953.6
ENST00000340870.6 |
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chr20_+_142573 | 0.06 |
ENST00000382398.4
|
DEFB126
|
defensin beta 126 |
| chr6_+_133889105 | 0.06 |
ENST00000367882.5
|
TCF21
|
transcription factor 21 |
| chr3_-_9769910 | 0.06 |
ENST00000256460.8
|
CAMK1
|
calcium/calmodulin dependent protein kinase I |
| chr10_+_97584314 | 0.06 |
ENST00000370647.8
|
HOGA1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr2_+_201132928 | 0.05 |
ENST00000462763.5
|
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chr6_-_87095059 | 0.05 |
ENST00000369582.6
ENST00000610310.3 ENST00000630630.2 ENST00000627148.3 ENST00000625577.1 |
CGA
|
glycoprotein hormones, alpha polypeptide |
| chr11_+_67403887 | 0.05 |
ENST00000526387.5
|
TBC1D10C
|
TBC1 domain family member 10C |
| chr2_+_227813834 | 0.05 |
ENST00000358813.5
ENST00000409189.7 |
CCL20
|
C-C motif chemokine ligand 20 |
| chr17_-_66220630 | 0.05 |
ENST00000585162.1
|
APOH
|
apolipoprotein H |
| chr11_+_67404077 | 0.05 |
ENST00000542590.2
ENST00000312390.9 |
TBC1D10C
|
TBC1 domain family member 10C |
| chr1_+_1281224 | 0.05 |
ENST00000400928.7
|
SCNN1D
|
sodium channel epithelial 1 subunit delta |
| chr20_-_57710539 | 0.05 |
ENST00000395816.7
ENST00000347215.8 |
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr19_-_12834548 | 0.05 |
ENST00000590404.6
ENST00000592204.5 ENST00000674343.2 |
RTBDN
|
retbindin |
| chr19_-_10102659 | 0.05 |
ENST00000592641.5
ENST00000253109.5 |
ANGPTL6
|
angiopoietin like 6 |
| chr2_-_196926717 | 0.05 |
ENST00000409475.5
ENST00000374738.3 |
PGAP1
|
post-GPI attachment to proteins inositol deacylase 1 |
| chr14_+_93207229 | 0.04 |
ENST00000554232.5
ENST00000556871.5 ENST00000013070.11 ENST00000555113.5 |
UBR7
|
ubiquitin protein ligase E3 component n-recognin 7 |
| chr14_-_74426102 | 0.04 |
ENST00000554953.1
ENST00000331628.8 |
SYNDIG1L
|
synapse differentiation inducing 1 like |
| chr12_+_25958891 | 0.04 |
ENST00000381352.7
ENST00000535907.5 ENST00000405154.6 ENST00000615708.4 |
RASSF8
|
Ras association domain family member 8 |
| chr5_-_41870416 | 0.04 |
ENST00000196371.10
|
OXCT1
|
3-oxoacid CoA-transferase 1 |
| chr7_-_117323041 | 0.04 |
ENST00000491214.1
ENST00000265441.8 |
WNT2
|
Wnt family member 2 |
| chr16_-_89490479 | 0.04 |
ENST00000642600.1
ENST00000301030.10 |
ANKRD11
|
ankyrin repeat domain 11 |
| chr3_+_112333147 | 0.04 |
ENST00000473539.5
ENST00000315711.12 ENST00000383681.7 |
CD200
|
CD200 molecule |
| chr19_-_12835400 | 0.04 |
ENST00000591512.2
ENST00000587549.1 ENST00000322912.9 |
RTBDN
|
retbindin |
| chr19_-_12835421 | 0.04 |
ENST00000589272.5
|
RTBDN
|
retbindin |
| chr12_-_7109176 | 0.04 |
ENST00000545280.5
ENST00000543933.5 ENST00000545337.1 ENST00000266542.9 ENST00000544702.5 |
C1RL
|
complement C1r subcomponent like |
| chr2_+_201132769 | 0.04 |
ENST00000494258.5
|
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chr19_+_7920313 | 0.04 |
ENST00000221573.11
ENST00000595637.1 |
SNAPC2
|
small nuclear RNA activating complex polypeptide 2 |
| chr3_-_15341368 | 0.04 |
ENST00000408919.7
|
SH3BP5
|
SH3 domain binding protein 5 |
| chr2_+_230996115 | 0.04 |
ENST00000424440.5
ENST00000452881.5 ENST00000433428.6 ENST00000455816.1 ENST00000440792.5 ENST00000423134.1 |
SPATA3
|
spermatogenesis associated 3 |
| chr17_-_8210203 | 0.03 |
ENST00000578549.5
ENST00000582368.5 |
AURKB
|
aurora kinase B |
| chr11_-_26722051 | 0.03 |
ENST00000396005.8
|
SLC5A12
|
solute carrier family 5 member 12 |
| chr18_+_7567266 | 0.03 |
ENST00000580170.6
|
PTPRM
|
protein tyrosine phosphatase receptor type M |
| chr15_-_78811415 | 0.03 |
ENST00000388820.5
|
ADAMTS7
|
ADAM metallopeptidase with thrombospondin type 1 motif 7 |
| chr15_-_60397964 | 0.03 |
ENST00000558998.5
ENST00000560165.5 ENST00000557986.5 ENST00000559467.5 ENST00000677968.1 ENST00000678450.1 ENST00000332680.8 ENST00000396024.7 ENST00000557906.6 ENST00000558558.6 ENST00000559113.6 ENST00000559780.6 ENST00000559956.6 ENST00000560468.6 ENST00000678870.1 ENST00000678061.1 ENST00000451270.7 ENST00000421017.6 ENST00000560466.5 ENST00000558132.5 ENST00000559370.5 ENST00000559725.5 ENST00000558985.6 ENST00000679109.1 |
ANXA2
|
annexin A2 |
| chr5_+_168486462 | 0.03 |
ENST00000231572.8
ENST00000626454.1 |
RARS1
|
arginyl-tRNA synthetase 1 |
| chr9_-_35828579 | 0.03 |
ENST00000377984.2
ENST00000423537.7 ENST00000472182.1 |
FAM221B
|
family with sequence similarity 221 member B |
| chr2_+_201132872 | 0.03 |
ENST00000470178.6
|
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chr14_-_93207007 | 0.03 |
ENST00000556566.1
ENST00000306954.5 |
GON7
|
GON7 subunit of KEOPS complex |
| chr6_+_123804141 | 0.03 |
ENST00000368416.5
|
NKAIN2
|
sodium/potassium transporting ATPase interacting 2 |
| chr11_-_6771976 | 0.03 |
ENST00000641124.1
ENST00000641169.1 |
OR2AG2
|
olfactory receptor family 2 subfamily AG member 2 |
| chrX_+_71144818 | 0.03 |
ENST00000536169.5
ENST00000358741.4 ENST00000395855.6 ENST00000374051.7 |
NLGN3
|
neuroligin 3 |
| chr12_-_53336342 | 0.03 |
ENST00000537210.2
ENST00000536324.4 |
SP7
|
Sp7 transcription factor |
| chr9_+_70043840 | 0.03 |
ENST00000377182.5
|
MAMDC2
|
MAM domain containing 2 |
| chr3_+_15601736 | 0.03 |
ENST00000303498.10
ENST00000417015.3 ENST00000672065.1 ENST00000672112.1 ENST00000643237.3 ENST00000672141.1 ENST00000646371.1 ENST00000672427.1 ENST00000672336.1 ENST00000673467.1 ENST00000672760.1 ENST00000437172.6 |
BTD
|
biotinidase |
| chr19_+_4304588 | 0.03 |
ENST00000221856.11
|
FSD1
|
fibronectin type III and SPRY domain containing 1 |
| chr12_+_109900258 | 0.02 |
ENST00000405876.9
|
TCHP
|
trichoplein keratin filament binding |
| chr22_-_23751080 | 0.02 |
ENST00000341976.5
|
ZNF70
|
zinc finger protein 70 |
| chr10_+_116590956 | 0.02 |
ENST00000358834.9
ENST00000528052.5 |
PNLIPRP1
|
pancreatic lipase related protein 1 |
| chr19_+_55488404 | 0.02 |
ENST00000594321.5
ENST00000389623.11 |
SSC5D
|
scavenger receptor cysteine rich family member with 5 domains |
| chr12_-_53335737 | 0.02 |
ENST00000303846.3
|
SP7
|
Sp7 transcription factor |
| chr12_+_120694167 | 0.02 |
ENST00000535656.1
|
MLEC
|
malectin |
| chr16_+_6019663 | 0.02 |
ENST00000422070.8
|
RBFOX1
|
RNA binding fox-1 homolog 1 |
| chr22_+_24607638 | 0.02 |
ENST00000432867.5
|
GGT1
|
gamma-glutamyltransferase 1 |
| chr14_+_21030509 | 0.02 |
ENST00000481535.5
|
TPPP2
|
tubulin polymerization promoting protein family member 2 |
| chr17_-_63704388 | 0.02 |
ENST00000582026.2
|
STRADA
|
STE20 related adaptor alpha |
| chr19_-_37210484 | 0.02 |
ENST00000527838.5
ENST00000591492.5 ENST00000532828.7 |
ZNF585B
|
zinc finger protein 585B |
| chr19_-_40425982 | 0.02 |
ENST00000357949.5
|
SERTAD1
|
SERTA domain containing 1 |
| chr2_-_196926670 | 0.02 |
ENST00000354764.9
|
PGAP1
|
post-GPI attachment to proteins inositol deacylase 1 |
| chr10_+_116591010 | 0.02 |
ENST00000530319.5
ENST00000527980.5 ENST00000471549.5 ENST00000534537.5 |
PNLIPRP1
|
pancreatic lipase related protein 1 |
| chr17_+_47831608 | 0.02 |
ENST00000269025.9
|
LRRC46
|
leucine rich repeat containing 46 |
| chr1_-_155934398 | 0.02 |
ENST00000368320.7
ENST00000368321.8 |
KHDC4
|
KH domain containing 4, pre-mRNA splicing factor |
| chr6_-_111873421 | 0.02 |
ENST00000368678.8
ENST00000523238.5 ENST00000354650.7 |
FYN
|
FYN proto-oncogene, Src family tyrosine kinase |
| chr11_+_2302119 | 0.02 |
ENST00000381121.7
|
TSPAN32
|
tetraspanin 32 |
| chr7_-_141014939 | 0.02 |
ENST00000324787.10
ENST00000467334.1 |
MRPS33
|
mitochondrial ribosomal protein S33 |
| chr5_-_181205284 | 0.02 |
ENST00000334421.5
|
TRIM7
|
tripartite motif containing 7 |
| chr14_+_73616844 | 0.02 |
ENST00000381139.1
|
ACOT6
|
acyl-CoA thioesterase 6 |
| chr4_-_25160375 | 0.02 |
ENST00000382103.7
|
SEPSECS
|
Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase |
| chr5_+_144205250 | 0.01 |
ENST00000507359.3
|
KCTD16
|
potassium channel tetramerization domain containing 16 |
| chr12_+_53454718 | 0.01 |
ENST00000552819.5
ENST00000455667.7 |
PCBP2
|
poly(rC) binding protein 2 |
| chr10_+_50990864 | 0.01 |
ENST00000401604.8
|
PRKG1
|
protein kinase cGMP-dependent 1 |
| chr4_-_25160546 | 0.01 |
ENST00000681948.1
|
SEPSECS
|
Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase |
| chr10_+_101354083 | 0.01 |
ENST00000408038.6
ENST00000370187.8 |
BTRC
|
beta-transducin repeat containing E3 ubiquitin protein ligase |
| chr1_+_61203496 | 0.01 |
ENST00000663597.1
|
NFIA
|
nuclear factor I A |
| chr17_-_47831509 | 0.01 |
ENST00000414011.1
ENST00000351111.7 |
MRPL10
|
mitochondrial ribosomal protein L10 |
| chr12_-_56360084 | 0.01 |
ENST00000314128.9
ENST00000557235.5 ENST00000651915.1 |
STAT2
|
signal transducer and activator of transcription 2 |
| chr1_+_202122881 | 0.01 |
ENST00000683302.1
ENST00000683557.1 |
GPR37L1
|
G protein-coupled receptor 37 like 1 |
| chr2_-_42361198 | 0.01 |
ENST00000234301.3
|
COX7A2L
|
cytochrome c oxidase subunit 7A2 like |
| chr21_+_33230073 | 0.01 |
ENST00000342101.7
ENST00000413881.5 ENST00000443073.5 |
IFNAR2
|
interferon alpha and beta receptor subunit 2 |
| chr6_-_117602447 | 0.01 |
ENST00000368498.7
ENST00000052569.10 ENST00000535237.2 |
GOPC
|
golgi associated PDZ and coiled-coil motif containing |
| chr17_+_6444441 | 0.01 |
ENST00000250056.12
ENST00000571373.5 ENST00000570337.6 ENST00000572595.6 ENST00000572447.6 ENST00000576056.5 |
PIMREG
|
PICALM interacting mitotic regulator |
| chr10_+_5048748 | 0.01 |
ENST00000602997.5
ENST00000439082.7 |
AKR1C3
|
aldo-keto reductase family 1 member C3 |
| chr6_-_36986122 | 0.01 |
ENST00000460219.2
ENST00000373627.10 ENST00000373616.9 |
MTCH1
|
mitochondrial carrier 1 |
| chr1_+_202122910 | 0.01 |
ENST00000682545.1
ENST00000367282.6 ENST00000682887.1 |
GPR37L1
|
G protein-coupled receptor 37 like 1 |
| chr4_-_40630588 | 0.01 |
ENST00000514014.1
|
RBM47
|
RNA binding motif protein 47 |
| chr1_+_6206077 | 0.01 |
ENST00000377939.5
|
RNF207
|
ring finger protein 207 |
| chr9_+_77177511 | 0.01 |
ENST00000360280.8
ENST00000645632.1 ENST00000643348.1 |
VPS13A
|
vacuolar protein sorting 13 homolog A |
| chr6_+_6588082 | 0.01 |
ENST00000379953.6
|
LY86
|
lymphocyte antigen 86 |
| chr8_+_93794816 | 0.01 |
ENST00000519845.5
ENST00000684343.1 |
TMEM67
|
transmembrane protein 67 |
| chr19_+_17075767 | 0.01 |
ENST00000682292.1
ENST00000595618.5 ENST00000594824.5 |
MYO9B
|
myosin IXB |
| chr6_+_6588708 | 0.01 |
ENST00000230568.5
|
LY86
|
lymphocyte antigen 86 |
| chr17_-_42028662 | 0.01 |
ENST00000461831.1
|
ZNF385C
|
zinc finger protein 385C |
| chr7_-_142258027 | 0.01 |
ENST00000552471.1
ENST00000547058.6 |
PRSS58
|
serine protease 58 |
| chr20_-_3786677 | 0.01 |
ENST00000379751.5
|
CENPB
|
centromere protein B |
| chr13_-_41019289 | 0.01 |
ENST00000239882.7
|
ELF1
|
E74 like ETS transcription factor 1 |
| chr1_-_51878711 | 0.01 |
ENST00000352171.12
|
NRDC
|
nardilysin convertase |
| chr18_-_46757012 | 0.01 |
ENST00000315087.12
|
ST8SIA5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr10_-_116273606 | 0.01 |
ENST00000682743.1
|
GFRA1
|
GDNF family receptor alpha 1 |
| chr18_-_46756791 | 0.01 |
ENST00000538168.5
ENST00000536490.1 |
ST8SIA5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr12_+_53454819 | 0.01 |
ENST00000562264.5
|
PCBP2
|
poly(rC) binding protein 2 |
| chr11_-_2301859 | 0.01 |
ENST00000456145.2
ENST00000381153.8 |
C11orf21
|
chromosome 11 open reading frame 21 |
| chr7_+_100219236 | 0.01 |
ENST00000317271.2
|
PVRIG
|
PVR related immunoglobulin domain containing |
| chr12_+_67648737 | 0.01 |
ENST00000344096.4
ENST00000393555.3 |
DYRK2
|
dual specificity tyrosine phosphorylation regulated kinase 2 |
| chr10_+_101354028 | 0.01 |
ENST00000393441.8
|
BTRC
|
beta-transducin repeat containing E3 ubiquitin protein ligase |
| chr6_-_52803807 | 0.00 |
ENST00000334575.6
|
GSTA1
|
glutathione S-transferase alpha 1 |
| chr3_+_46877705 | 0.00 |
ENST00000449590.6
|
PTH1R
|
parathyroid hormone 1 receptor |
| chr1_-_51878799 | 0.00 |
ENST00000354831.11
ENST00000544028.5 |
NRDC
|
nardilysin convertase |
| chr21_+_33229883 | 0.00 |
ENST00000382264.7
ENST00000342136.9 ENST00000404220.7 |
IFNAR2
|
interferon alpha and beta receptor subunit 2 |
| chr16_+_56935371 | 0.00 |
ENST00000568358.1
|
HERPUD1
|
homocysteine inducible ER protein with ubiquitin like domain 1 |
| chr12_-_68253502 | 0.00 |
ENST00000328087.6
ENST00000538666.6 |
IL22
|
interleukin 22 |
| chr12_+_53454764 | 0.00 |
ENST00000439930.7
ENST00000548933.5 |
PCBP2
|
poly(rC) binding protein 2 |
| chr1_+_45012896 | 0.00 |
ENST00000428106.1
|
UROD
|
uroporphyrinogen decarboxylase |
| chr1_-_10947870 | 0.00 |
ENST00000468348.1
|
C1orf127
|
chromosome 1 open reading frame 127 |
| chr12_+_49295138 | 0.00 |
ENST00000257860.9
|
PRPH
|
peripherin |
| chr2_-_128027273 | 0.00 |
ENST00000259235.7
ENST00000357702.9 ENST00000424298.5 |
SAP130
|
Sin3A associated protein 130 |
| chr14_+_104138578 | 0.00 |
ENST00000423312.7
|
KIF26A
|
kinesin family member 26A |
| chr14_+_23953729 | 0.00 |
ENST00000558263.5
ENST00000543741.6 ENST00000313250.10 ENST00000397075.7 ENST00000397074.7 ENST00000559632.5 ENST00000558581.5 |
DHRS4
|
dehydrogenase/reductase 4 |
| chr1_-_53940100 | 0.00 |
ENST00000371376.1
|
HSPB11
|
heat shock protein family B (small) member 11 |
| chr4_+_25160631 | 0.00 |
ENST00000510415.1
ENST00000507794.2 ENST00000512921.4 |
SEPSECS-AS1
PI4K2B
|
SEPSECS antisense RNA 1 (head to head) phosphatidylinositol 4-kinase type 2 beta |
| chr1_+_45012691 | 0.00 |
ENST00000469548.5
|
UROD
|
uroporphyrinogen decarboxylase |
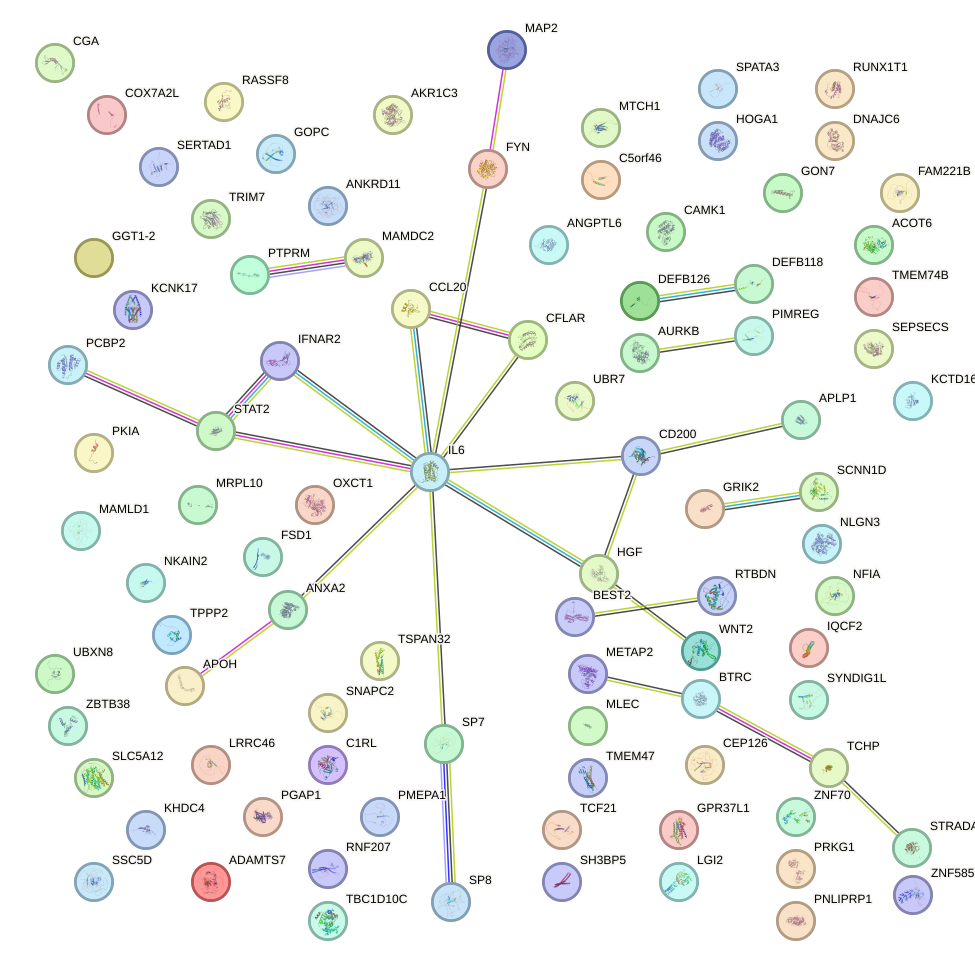
Network of associatons between targets according to the STRING database.
First level regulatory network of HAND1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.2 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.1 | GO:0072276 | branchiomeric skeletal muscle development(GO:0014707) bronchiole development(GO:0060435) metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.0 | 0.1 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.0 | 0.2 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.1 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.0 | 0.1 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.0 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 0.2 | GO:0031696 | alpha-2C adrenergic receptor binding(GO:0031696) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:1902444 | riboflavin binding(GO:1902444) |
| 0.0 | 0.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |