Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
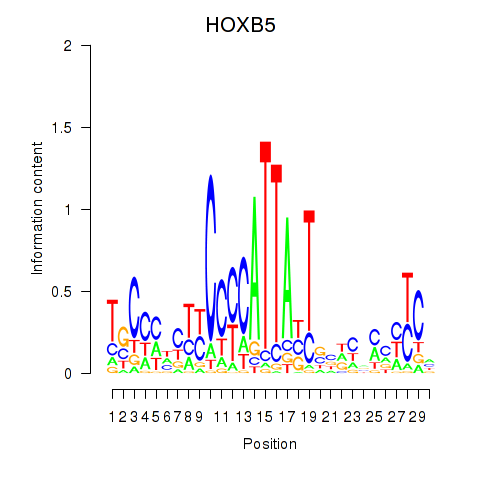
Results for HOXB5
Z-value: 0.20
Transcription factors associated with HOXB5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB5
|
ENSG00000120075.6 | homeobox B5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB5 | hg38_v1_chr17_-_48593748_48593786 | -0.15 | 7.3e-01 | Click! |
Activity profile of HOXB5 motif
Sorted Z-values of HOXB5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_4654732 | 0.19 |
ENST00000378190.7
|
AJAP1
|
adherens junctions associated protein 1 |
| chr19_-_15934410 | 0.17 |
ENST00000326742.12
|
CYP4F11
|
cytochrome P450 family 4 subfamily F member 11 |
| chr11_-_125778818 | 0.16 |
ENST00000358524.8
|
PATE2
|
prostate and testis expressed 2 |
| chr11_-_125778788 | 0.16 |
ENST00000436890.2
|
PATE2
|
prostate and testis expressed 2 |
| chr19_-_15934521 | 0.15 |
ENST00000402119.9
|
CYP4F11
|
cytochrome P450 family 4 subfamily F member 11 |
| chr2_+_17541157 | 0.12 |
ENST00000406397.1
|
VSNL1
|
visinin like 1 |
| chr6_-_96837460 | 0.12 |
ENST00000229955.4
|
GPR63
|
G protein-coupled receptor 63 |
| chr2_+_17540670 | 0.12 |
ENST00000451533.5
ENST00000295156.9 |
VSNL1
|
visinin like 1 |
| chr5_-_35047935 | 0.11 |
ENST00000510428.1
ENST00000231420.11 |
AGXT2
|
alanine--glyoxylate aminotransferase 2 |
| chr1_+_4654601 | 0.09 |
ENST00000378191.5
|
AJAP1
|
adherens junctions associated protein 1 |
| chr15_-_74725370 | 0.07 |
ENST00000567032.5
ENST00000564596.5 ENST00000566503.1 ENST00000395049.8 ENST00000379727.8 ENST00000617691.4 ENST00000395048.6 |
CYP1A1
|
cytochrome P450 family 1 subfamily A member 1 |
| chr12_+_57591158 | 0.06 |
ENST00000422156.7
ENST00000354947.10 ENST00000540759.6 ENST00000551772.5 ENST00000550465.5 |
PIP4K2C
|
phosphatidylinositol-5-phosphate 4-kinase type 2 gamma |
| chr3_+_45945019 | 0.06 |
ENST00000458629.1
ENST00000457814.1 |
CXCR6
|
C-X-C motif chemokine receptor 6 |
| chr14_+_21030201 | 0.05 |
ENST00000321760.11
ENST00000460647.6 ENST00000530140.6 ENST00000472458.5 |
TPPP2
|
tubulin polymerization promoting protein family member 2 |
| chr15_-_48811038 | 0.05 |
ENST00000380950.7
|
CEP152
|
centrosomal protein 152 |
| chr1_+_116373233 | 0.04 |
ENST00000295598.10
|
ATP1A1
|
ATPase Na+/K+ transporting subunit alpha 1 |
| chrX_+_48521817 | 0.04 |
ENST00000446158.5
ENST00000414061.1 |
EBP
|
EBP cholestenol delta-isomerase |
| chrX_+_48521788 | 0.04 |
ENST00000651615.1
ENST00000495186.6 |
ENSG00000286268.1
EBP
|
novel protein EBP cholestenol delta-isomerase |
| chr12_-_54981838 | 0.04 |
ENST00000316577.12
|
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr20_+_8789517 | 0.04 |
ENST00000437439.2
|
PLCB1
|
phospholipase C beta 1 |
| chr1_+_157993601 | 0.04 |
ENST00000359209.11
|
KIRREL1
|
kirre like nephrin family adhesion molecule 1 |
| chr11_-_6030758 | 0.04 |
ENST00000641900.1
|
OR56A1
|
olfactory receptor family 56 subfamily A member 1 |
| chr3_+_130894382 | 0.04 |
ENST00000509662.5
ENST00000328560.12 ENST00000428331.6 ENST00000359644.7 ENST00000422190.6 |
ATP2C1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr12_+_54561442 | 0.03 |
ENST00000550620.1
|
PDE1B
|
phosphodiesterase 1B |
| chr16_-_84618067 | 0.03 |
ENST00000262428.5
|
COTL1
|
coactosin like F-actin binding protein 1 |
| chr16_-_84618041 | 0.03 |
ENST00000564057.1
|
COTL1
|
coactosin like F-actin binding protein 1 |
| chr11_-_57322197 | 0.03 |
ENST00000532437.1
|
TNKS1BP1
|
tankyrase 1 binding protein 1 |
| chr6_+_26021641 | 0.03 |
ENST00000617569.2
|
H4C1
|
H4 clustered histone 1 |
| chr6_-_31541937 | 0.03 |
ENST00000456662.5
ENST00000431908.5 ENST00000456976.5 ENST00000428450.5 ENST00000418897.5 ENST00000396172.6 ENST00000419020.1 ENST00000428098.5 |
DDX39B
|
DExD-box helicase 39B |
| chr3_+_51385282 | 0.03 |
ENST00000528157.7
|
MANF
|
mesencephalic astrocyte derived neurotrophic factor |
| chr10_-_101843777 | 0.03 |
ENST00000356640.7
|
KCNIP2
|
potassium voltage-gated channel interacting protein 2 |
| chr8_-_92095598 | 0.03 |
ENST00000520724.5
ENST00000518844.5 |
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1 |
| chr8_-_92095627 | 0.03 |
ENST00000517919.5
ENST00000617740.4 ENST00000613302.4 ENST00000436581.6 ENST00000614812.4 ENST00000519847.5 |
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1 |
| chr9_-_127937800 | 0.02 |
ENST00000373110.4
ENST00000314392.13 |
DPM2
|
dolichyl-phosphate mannosyltransferase subunit 2, regulatory |
| chrX_+_14529516 | 0.02 |
ENST00000218075.9
|
GLRA2
|
glycine receptor alpha 2 |
| chr19_+_13151975 | 0.02 |
ENST00000588173.1
|
IER2
|
immediate early response 2 |
| chr21_-_44262216 | 0.02 |
ENST00000270172.7
|
DNMT3L
|
DNA methyltransferase 3 like |
| chr18_-_35497591 | 0.02 |
ENST00000589273.1
ENST00000586489.5 |
INO80C
|
INO80 complex subunit C |
| chr11_+_109422174 | 0.02 |
ENST00000327419.7
|
C11orf87
|
chromosome 11 open reading frame 87 |
| chr17_-_17237122 | 0.02 |
ENST00000389169.9
ENST00000285071.9 ENST00000417064.1 |
FLCN
|
folliculin |
| chr2_-_219308963 | 0.02 |
ENST00000423636.6
ENST00000442029.5 ENST00000412847.5 |
PTPRN
|
protein tyrosine phosphatase receptor type N |
| chr11_+_4643966 | 0.02 |
ENST00000396952.6
|
OR51E1
|
olfactory receptor family 51 subfamily E member 1 |
| chr8_-_92095215 | 0.02 |
ENST00000360348.6
ENST00000520428.5 ENST00000518992.5 ENST00000520556.5 ENST00000518317.5 ENST00000521319.5 ENST00000521375.5 ENST00000518449.5 ENST00000613886.4 |
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1 |
| chr7_+_130381092 | 0.02 |
ENST00000484324.1
|
CPA1
|
carboxypeptidase A1 |
| chr11_-_128522189 | 0.02 |
ENST00000526145.6
|
ETS1
|
ETS proto-oncogene 1, transcription factor |
| chr6_-_31542339 | 0.02 |
ENST00000458640.5
|
DDX39B
|
DExD-box helicase 39B |
| chr8_+_117135259 | 0.02 |
ENST00000519688.5
|
SLC30A8
|
solute carrier family 30 member 8 |
| chr8_-_132085655 | 0.02 |
ENST00000262283.5
|
ENSG00000258417.3
|
novel protein |
| chr20_-_49568101 | 0.02 |
ENST00000244043.5
|
PTGIS
|
prostaglandin I2 synthase |
| chr16_-_57971086 | 0.02 |
ENST00000564448.5
ENST00000311183.8 |
CNGB1
|
cyclic nucleotide gated channel subunit beta 1 |
| chr20_-_6054026 | 0.02 |
ENST00000378858.5
|
LRRN4
|
leucine rich repeat neuronal 4 |
| chr3_-_195811889 | 0.02 |
ENST00000475231.5
|
MUC4
|
mucin 4, cell surface associated |
| chr9_+_4662281 | 0.02 |
ENST00000381883.5
|
PLPP6
|
phospholipid phosphatase 6 |
| chr14_+_21030509 | 0.02 |
ENST00000481535.5
|
TPPP2
|
tubulin polymerization promoting protein family member 2 |
| chr3_-_195811916 | 0.01 |
ENST00000463781.8
|
MUC4
|
mucin 4, cell surface associated |
| chr3_-_195811857 | 0.01 |
ENST00000349607.8
ENST00000346145.8 |
MUC4
|
mucin 4, cell surface associated |
| chrX_+_101078861 | 0.01 |
ENST00000372930.5
|
TMEM35A
|
transmembrane protein 35A |
| chr6_+_167111789 | 0.01 |
ENST00000400926.5
|
CCR6
|
C-C motif chemokine receptor 6 |
| chr1_-_36385887 | 0.01 |
ENST00000373130.7
ENST00000373132.4 |
STK40
|
serine/threonine kinase 40 |
| chr1_-_36385872 | 0.01 |
ENST00000373129.7
|
STK40
|
serine/threonine kinase 40 |
| chr10_+_112376193 | 0.01 |
ENST00000433418.6
ENST00000356116.6 ENST00000354273.5 |
ACSL5
|
acyl-CoA synthetase long chain family member 5 |
| chr19_-_42220109 | 0.01 |
ENST00000595337.5
|
DEDD2
|
death effector domain containing 2 |
| chr1_+_40691998 | 0.01 |
ENST00000534399.5
ENST00000372653.5 |
NFYC
|
nuclear transcription factor Y subunit gamma |
| chr18_+_34978244 | 0.01 |
ENST00000436190.6
|
MAPRE2
|
microtubule associated protein RP/EB family member 2 |
| chr5_-_161546970 | 0.01 |
ENST00000675303.1
|
GABRB2
|
gamma-aminobutyric acid type A receptor subunit beta2 |
| chr7_+_5282935 | 0.01 |
ENST00000396872.8
ENST00000444741.5 ENST00000297195.8 ENST00000406453.3 |
SLC29A4
|
solute carrier family 29 member 4 |
| chr16_-_57971121 | 0.01 |
ENST00000251102.13
|
CNGB1
|
cyclic nucleotide gated channel subunit beta 1 |
| chr19_-_49763295 | 0.01 |
ENST00000246801.8
|
TSKS
|
testis specific serine kinase substrate |
| chr12_-_6689450 | 0.01 |
ENST00000355772.8
ENST00000417772.7 ENST00000319770.7 ENST00000396801.7 |
ZNF384
|
zinc finger protein 384 |
| chr17_-_30906202 | 0.00 |
ENST00000580840.1
ENST00000581216.6 |
TEFM
|
transcription elongation factor, mitochondrial |
| chr12_-_6689359 | 0.00 |
ENST00000683879.1
|
ZNF384
|
zinc finger protein 384 |
| chr1_+_40691749 | 0.00 |
ENST00000372654.5
|
NFYC
|
nuclear transcription factor Y subunit gamma |
| chr1_+_40691689 | 0.00 |
ENST00000427410.6
ENST00000447388.7 ENST00000425457.6 ENST00000453631.5 ENST00000456393.6 |
NFYC
|
nuclear transcription factor Y subunit gamma |
| chr1_+_162381703 | 0.00 |
ENST00000458626.4
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr14_-_23551729 | 0.00 |
ENST00000419474.5
|
ZFHX2
|
zinc finger homeobox 2 |
| chr12_-_6689244 | 0.00 |
ENST00000361959.7
ENST00000436774.6 ENST00000544482.1 |
ZNF384
|
zinc finger protein 384 |
| chr17_-_7207245 | 0.00 |
ENST00000649971.1
|
DLG4
|
discs large MAGUK scaffold protein 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0042377 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.0 | 0.3 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.0 | 0.1 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.0 | 0.1 | GO:0043449 | cellular alkene metabolic process(GO:0043449) |
| 0.0 | 0.1 | GO:0033490 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) cholestenol delta-isomerase activity(GO:0047750) |
| 0.0 | 0.1 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |