Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
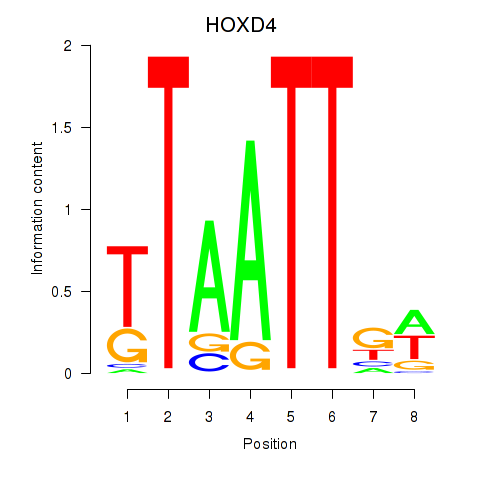
Results for HOXD4
Z-value: 0.50
Transcription factors associated with HOXD4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXD4
|
ENSG00000170166.6 | homeobox D4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXD4 | hg38_v1_chr2_+_176151543_176151559 | -0.10 | 8.1e-01 | Click! |
Activity profile of HOXD4 motif
Sorted Z-values of HOXD4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_31447732 | 0.96 |
ENST00000257189.5
|
DSG3
|
desmoglein 3 |
| chr2_-_70553638 | 0.50 |
ENST00000444975.5
ENST00000445399.5 ENST00000295400.11 ENST00000418333.6 |
TGFA
|
transforming growth factor alpha |
| chr6_+_130018565 | 0.41 |
ENST00000361794.7
ENST00000526087.5 ENST00000533560.5 |
L3MBTL3
|
L3MBTL histone methyl-lysine binding protein 3 |
| chr6_-_136526472 | 0.35 |
ENST00000454590.5
ENST00000432797.6 |
MAP7
|
microtubule associated protein 7 |
| chr7_-_22193728 | 0.34 |
ENST00000620335.4
|
RAPGEF5
|
Rap guanine nucleotide exchange factor 5 |
| chr6_-_136526177 | 0.34 |
ENST00000617204.4
|
MAP7
|
microtubule associated protein 7 |
| chr7_-_22193824 | 0.32 |
ENST00000401957.6
|
RAPGEF5
|
Rap guanine nucleotide exchange factor 5 |
| chr12_-_39340963 | 0.30 |
ENST00000552961.5
|
KIF21A
|
kinesin family member 21A |
| chr2_-_70553440 | 0.30 |
ENST00000450929.5
|
TGFA
|
transforming growth factor alpha |
| chr5_+_140841183 | 0.29 |
ENST00000378123.4
ENST00000531613.2 |
PCDHA8
|
protocadherin alpha 8 |
| chr4_+_40197023 | 0.28 |
ENST00000381799.10
|
RHOH
|
ras homolog family member H |
| chr12_+_41437680 | 0.27 |
ENST00000649474.1
ENST00000539469.6 ENST00000298919.7 |
PDZRN4
|
PDZ domain containing ring finger 4 |
| chr5_+_67004618 | 0.23 |
ENST00000261569.11
ENST00000436277.5 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr11_-_72781858 | 0.21 |
ENST00000537947.5
|
STARD10
|
StAR related lipid transfer domain containing 10 |
| chr9_+_470291 | 0.21 |
ENST00000382303.5
|
KANK1
|
KN motif and ankyrin repeat domains 1 |
| chr4_-_142305826 | 0.20 |
ENST00000514525.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase type II B |
| chr4_-_142305935 | 0.20 |
ENST00000511838.5
|
INPP4B
|
inositol polyphosphate-4-phosphatase type II B |
| chr3_+_111998915 | 0.18 |
ENST00000478951.6
|
TAGLN3
|
transgelin 3 |
| chr17_-_10549652 | 0.18 |
ENST00000245503.10
|
MYH2
|
myosin heavy chain 2 |
| chr3_+_111999326 | 0.18 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr4_+_68447453 | 0.18 |
ENST00000305363.9
|
TMPRSS11E
|
transmembrane serine protease 11E |
| chr17_-_28406160 | 0.18 |
ENST00000618626.1
ENST00000612814.5 |
SLC46A1
|
solute carrier family 46 member 1 |
| chr17_-_10549694 | 0.18 |
ENST00000622564.4
|
MYH2
|
myosin heavy chain 2 |
| chr7_+_92057602 | 0.17 |
ENST00000491695.2
|
AKAP9
|
A-kinase anchoring protein 9 |
| chr17_-_10549612 | 0.17 |
ENST00000532183.6
ENST00000397183.6 ENST00000420805.1 |
MYH2
|
myosin heavy chain 2 |
| chr3_+_142623386 | 0.17 |
ENST00000337777.7
ENST00000497199.5 |
PLS1
|
plastin 1 |
| chr5_+_69415065 | 0.17 |
ENST00000647531.1
ENST00000645446.1 ENST00000325631.10 ENST00000454295.6 |
MARVELD2
|
MARVEL domain containing 2 |
| chr4_-_152679984 | 0.17 |
ENST00000304385.8
ENST00000504064.1 |
TMEM154
|
transmembrane protein 154 |
| chr17_-_66229380 | 0.16 |
ENST00000205948.11
|
APOH
|
apolipoprotein H |
| chr11_-_72781833 | 0.16 |
ENST00000535054.1
ENST00000545082.5 |
STARD10
|
StAR related lipid transfer domain containing 10 |
| chr1_+_186828941 | 0.15 |
ENST00000367466.4
|
PLA2G4A
|
phospholipase A2 group IVA |
| chr2_+_203936755 | 0.14 |
ENST00000316386.11
ENST00000435193.1 |
ICOS
|
inducible T cell costimulator |
| chr2_-_99255107 | 0.13 |
ENST00000333017.6
ENST00000626374.2 ENST00000409679.5 ENST00000423306.1 |
LYG2
|
lysozyme g2 |
| chr4_-_129093548 | 0.13 |
ENST00000503401.1
|
SCLT1
|
sodium channel and clathrin linker 1 |
| chr17_-_40782544 | 0.13 |
ENST00000301656.4
|
KRT27
|
keratin 27 |
| chr1_-_152115443 | 0.12 |
ENST00000614923.1
|
TCHH
|
trichohyalin |
| chr18_-_21703688 | 0.12 |
ENST00000584464.1
ENST00000578270.5 |
ABHD3
|
abhydrolase domain containing 3, phospholipase |
| chr7_-_137343752 | 0.12 |
ENST00000393083.2
|
PTN
|
pleiotrophin |
| chr4_-_76023489 | 0.11 |
ENST00000306602.3
|
CXCL10
|
C-X-C motif chemokine ligand 10 |
| chr2_+_190180835 | 0.11 |
ENST00000340623.4
|
C2orf88
|
chromosome 2 open reading frame 88 |
| chr2_+_71453538 | 0.11 |
ENST00000258104.8
|
DYSF
|
dysferlin |
| chr11_+_59436469 | 0.11 |
ENST00000641045.1
|
OR5A1
|
olfactory receptor family 5 subfamily A member 1 |
| chr6_-_53148822 | 0.10 |
ENST00000259803.8
|
GCM1
|
glial cells missing transcription factor 1 |
| chr10_-_49762276 | 0.10 |
ENST00000374103.9
|
OGDHL
|
oxoglutarate dehydrogenase L |
| chr2_+_108377947 | 0.10 |
ENST00000272452.7
|
SULT1C4
|
sulfotransferase family 1C member 4 |
| chr10_-_49762335 | 0.10 |
ENST00000419399.4
ENST00000432695.2 |
OGDHL
|
oxoglutarate dehydrogenase L |
| chr2_-_207167220 | 0.10 |
ENST00000421199.5
ENST00000457962.5 |
KLF7
|
Kruppel like factor 7 |
| chr20_+_8789517 | 0.09 |
ENST00000437439.2
|
PLCB1
|
phospholipase C beta 1 |
| chr2_+_32277883 | 0.09 |
ENST00000238831.9
|
YIPF4
|
Yip1 domain family member 4 |
| chr1_-_9751540 | 0.09 |
ENST00000435891.5
|
CLSTN1
|
calsyntenin 1 |
| chr1_+_182450132 | 0.09 |
ENST00000294854.13
|
RGSL1
|
regulator of G protein signaling like 1 |
| chr15_-_44711306 | 0.09 |
ENST00000682850.1
|
PATL2
|
PAT1 homolog 2 |
| chr7_-_25228485 | 0.09 |
ENST00000222674.2
|
NPVF
|
neuropeptide VF precursor |
| chr17_+_21126947 | 0.09 |
ENST00000579303.5
|
DHRS7B
|
dehydrogenase/reductase 7B |
| chr3_+_141262614 | 0.09 |
ENST00000504264.5
|
PXYLP1
|
2-phosphoxylose phosphatase 1 |
| chr12_+_112906777 | 0.08 |
ENST00000452357.7
ENST00000445409.7 |
OAS1
|
2'-5'-oligoadenylate synthetase 1 |
| chr21_+_42403856 | 0.08 |
ENST00000291535.11
|
UBASH3A
|
ubiquitin associated and SH3 domain containing A |
| chr3_+_109136707 | 0.08 |
ENST00000622536.6
|
C3orf85
|
chromosome 3 open reading frame 85 |
| chr8_+_49909783 | 0.08 |
ENST00000518864.5
|
SNTG1
|
syntrophin gamma 1 |
| chr7_-_137343688 | 0.08 |
ENST00000348225.7
|
PTN
|
pleiotrophin |
| chr1_-_86914319 | 0.08 |
ENST00000611507.4
ENST00000616787.4 |
SELENOF
|
selenoprotein F |
| chr20_+_31475278 | 0.08 |
ENST00000201979.3
|
REM1
|
RRAD and GEM like GTPase 1 |
| chr4_-_129093377 | 0.08 |
ENST00000506368.5
ENST00000439369.6 ENST00000503215.5 |
SCLT1
|
sodium channel and clathrin linker 1 |
| chr4_-_129093454 | 0.08 |
ENST00000281142.10
ENST00000511426.5 |
SCLT1
|
sodium channel and clathrin linker 1 |
| chr4_+_158210479 | 0.08 |
ENST00000504569.5
ENST00000509278.5 ENST00000514558.5 ENST00000503200.5 ENST00000296529.11 |
TMEM144
|
transmembrane protein 144 |
| chr4_+_127632926 | 0.07 |
ENST00000335251.11
|
INTU
|
inturned planar cell polarity protein |
| chr7_-_29146527 | 0.07 |
ENST00000265394.10
|
CPVL
|
carboxypeptidase vitellogenic like |
| chr5_-_35047935 | 0.07 |
ENST00000510428.1
ENST00000231420.11 |
AGXT2
|
alanine--glyoxylate aminotransferase 2 |
| chr11_-_129192198 | 0.07 |
ENST00000310343.13
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr6_+_122996227 | 0.07 |
ENST00000275162.10
|
CLVS2
|
clavesin 2 |
| chr7_-_29146436 | 0.07 |
ENST00000396276.7
|
CPVL
|
carboxypeptidase vitellogenic like |
| chr2_+_190180930 | 0.07 |
ENST00000443551.2
|
C2orf88
|
chromosome 2 open reading frame 88 |
| chr7_-_80919017 | 0.07 |
ENST00000265361.8
|
SEMA3C
|
semaphorin 3C |
| chr7_-_108240049 | 0.07 |
ENST00000379022.8
|
NRCAM
|
neuronal cell adhesion molecule |
| chr1_-_92486916 | 0.07 |
ENST00000294702.6
|
GFI1
|
growth factor independent 1 transcriptional repressor |
| chr8_-_56113982 | 0.07 |
ENST00000311923.1
|
MOS
|
MOS proto-oncogene, serine/threonine kinase |
| chr1_+_197268204 | 0.06 |
ENST00000535699.5
ENST00000538660.5 |
CRB1
|
crumbs cell polarity complex component 1 |
| chr16_-_21652598 | 0.06 |
ENST00000569602.1
ENST00000268389.6 |
IGSF6
|
immunoglobulin superfamily member 6 |
| chr1_-_86914102 | 0.06 |
ENST00000331835.10
ENST00000401030.4 ENST00000370554.5 |
SELENOF
|
selenoprotein F |
| chr3_+_63652663 | 0.06 |
ENST00000343837.8
ENST00000469440.5 |
SNTN
|
sentan, cilia apical structure protein |
| chr13_+_42781578 | 0.06 |
ENST00000313851.3
|
FAM216B
|
family with sequence similarity 216 member B |
| chr3_+_4680617 | 0.06 |
ENST00000648212.1
|
ITPR1
|
inositol 1,4,5-trisphosphate receptor type 1 |
| chr2_-_169573856 | 0.06 |
ENST00000453153.7
ENST00000445210.1 |
FASTKD1
|
FAST kinase domains 1 |
| chr11_-_129192291 | 0.06 |
ENST00000682385.1
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chrX_-_10677720 | 0.06 |
ENST00000453318.6
|
MID1
|
midline 1 |
| chr6_-_26108125 | 0.06 |
ENST00000338379.6
|
H1-6
|
H1.6 linker histone, cluster member |
| chr3_-_197260369 | 0.06 |
ENST00000658155.1
ENST00000453607.5 |
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr2_+_233917371 | 0.06 |
ENST00000324695.9
ENST00000433712.6 |
TRPM8
|
transient receptor potential cation channel subfamily M member 8 |
| chr17_-_40799939 | 0.06 |
ENST00000306658.8
|
KRT28
|
keratin 28 |
| chr5_-_179617581 | 0.05 |
ENST00000523449.5
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 |
| chr2_-_158380960 | 0.05 |
ENST00000409187.5
|
CCDC148
|
coiled-coil domain containing 148 |
| chr9_-_21368962 | 0.05 |
ENST00000610660.1
|
IFNA13
|
interferon alpha 13 |
| chr4_+_154563003 | 0.05 |
ENST00000302068.9
ENST00000509493.1 |
FGB
|
fibrinogen beta chain |
| chr1_+_42380772 | 0.05 |
ENST00000431473.4
ENST00000410070.6 |
RIMKLA
|
ribosomal modification protein rimK like family member A |
| chr16_+_24537693 | 0.05 |
ENST00000564314.5
ENST00000567686.5 |
RBBP6
|
RB binding protein 6, ubiquitin ligase |
| chr13_+_108629605 | 0.05 |
ENST00000457511.7
|
MYO16
|
myosin XVI |
| chr8_+_97644164 | 0.05 |
ENST00000336273.8
|
MTDH
|
metadherin |
| chr2_+_27663441 | 0.05 |
ENST00000326019.10
ENST00000613058.4 |
SLC4A1AP
|
solute carrier family 4 member 1 adaptor protein |
| chr1_+_115029823 | 0.05 |
ENST00000256592.3
|
TSHB
|
thyroid stimulating hormone subunit beta |
| chr21_+_42403874 | 0.05 |
ENST00000319294.11
ENST00000398367.1 |
UBASH3A
|
ubiquitin associated and SH3 domain containing A |
| chr2_+_186590022 | 0.05 |
ENST00000261023.8
ENST00000374907.7 |
ITGAV
|
integrin subunit alpha V |
| chr4_+_69931066 | 0.05 |
ENST00000246891.9
|
CSN1S1
|
casein alpha s1 |
| chr3_-_138132381 | 0.05 |
ENST00000236709.4
|
A4GNT
|
alpha-1,4-N-acetylglucosaminyltransferase |
| chr1_+_67166448 | 0.04 |
ENST00000347310.10
|
IL23R
|
interleukin 23 receptor |
| chr1_-_150765785 | 0.04 |
ENST00000680311.1
ENST00000681728.1 ENST00000680288.1 |
CTSS
|
cathepsin S |
| chr22_-_30246739 | 0.04 |
ENST00000403987.3
ENST00000249075.4 |
LIF
|
LIF interleukin 6 family cytokine |
| chr22_+_41092869 | 0.04 |
ENST00000674155.1
|
EP300
|
E1A binding protein p300 |
| chr1_-_150765735 | 0.04 |
ENST00000679898.1
ENST00000448301.7 ENST00000680664.1 ENST00000679512.1 ENST00000368985.8 ENST00000679582.1 |
CTSS
|
cathepsin S |
| chr12_-_118190510 | 0.04 |
ENST00000540561.5
ENST00000537952.1 ENST00000537822.1 |
TAOK3
|
TAO kinase 3 |
| chr7_+_112423137 | 0.04 |
ENST00000005558.8
ENST00000621379.4 |
IFRD1
|
interferon related developmental regulator 1 |
| chr4_-_39977836 | 0.04 |
ENST00000303538.13
ENST00000503396.5 |
PDS5A
|
PDS5 cohesin associated factor A |
| chr17_+_42980547 | 0.04 |
ENST00000361677.5
ENST00000589705.1 |
RUNDC1
|
RUN domain containing 1 |
| chr5_-_62403506 | 0.04 |
ENST00000680062.1
|
DIMT1
|
DIMT1 rRNA methyltransferase and ribosome maturation factor |
| chr20_+_33562306 | 0.04 |
ENST00000344201.7
|
CBFA2T2
|
CBFA2/RUNX1 partner transcriptional co-repressor 2 |
| chr6_-_49744434 | 0.04 |
ENST00000433368.6
ENST00000354620.4 |
CRISP3
|
cysteine rich secretory protein 3 |
| chr18_+_63702958 | 0.04 |
ENST00000544088.6
|
SERPINB11
|
serpin family B member 11 |
| chr2_-_224569782 | 0.04 |
ENST00000409096.5
|
CUL3
|
cullin 3 |
| chr12_-_14961610 | 0.04 |
ENST00000542276.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor beta |
| chr5_+_141421020 | 0.03 |
ENST00000622044.1
ENST00000398587.7 |
PCDHGA11
|
protocadherin gamma subfamily A, 11 |
| chr15_+_80072559 | 0.03 |
ENST00000560228.5
ENST00000559835.5 ENST00000559775.5 ENST00000558688.5 ENST00000560392.5 ENST00000560976.5 ENST00000558272.5 ENST00000558390.5 |
ZFAND6
|
zinc finger AN1-type containing 6 |
| chr21_-_29061351 | 0.03 |
ENST00000432178.5
|
CCT8
|
chaperonin containing TCP1 subunit 8 |
| chr19_-_47886308 | 0.03 |
ENST00000222002.4
|
SULT2A1
|
sulfotransferase family 2A member 1 |
| chr6_+_46793379 | 0.03 |
ENST00000230588.9
ENST00000611727.2 |
MEP1A
|
meprin A subunit alpha |
| chr1_+_86914616 | 0.03 |
ENST00000370550.10
ENST00000370551.8 |
HS2ST1
|
heparan sulfate 2-O-sulfotransferase 1 |
| chr3_-_114178698 | 0.03 |
ENST00000467632.5
|
DRD3
|
dopamine receptor D3 |
| chr6_+_143677935 | 0.03 |
ENST00000440869.6
ENST00000367582.7 ENST00000451827.6 |
PHACTR2
|
phosphatase and actin regulator 2 |
| chr8_+_7894674 | 0.03 |
ENST00000302247.3
|
DEFB4A
|
defensin beta 4A |
| chr9_+_273026 | 0.03 |
ENST00000682249.1
ENST00000453981.5 ENST00000487230.5 ENST00000469391.5 |
DOCK8
|
dedicator of cytokinesis 8 |
| chr12_-_100262356 | 0.03 |
ENST00000548313.5
|
DEPDC4
|
DEP domain containing 4 |
| chr20_+_33562365 | 0.03 |
ENST00000346541.7
ENST00000397800.5 ENST00000492345.5 |
CBFA2T2
|
CBFA2/RUNX1 partner transcriptional co-repressor 2 |
| chr6_-_49744378 | 0.03 |
ENST00000371159.8
ENST00000263045.9 |
CRISP3
|
cysteine rich secretory protein 3 |
| chr17_+_9576627 | 0.03 |
ENST00000396219.7
ENST00000352665.10 |
CFAP52
|
cilia and flagella associated protein 52 |
| chr12_+_14365661 | 0.03 |
ENST00000261168.9
ENST00000538511.5 ENST00000545723.1 ENST00000543189.5 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr12_-_48852153 | 0.03 |
ENST00000308025.8
|
DDX23
|
DEAD-box helicase 23 |
| chrX_+_56563569 | 0.03 |
ENST00000338222.7
|
UBQLN2
|
ubiquilin 2 |
| chr20_-_10420737 | 0.02 |
ENST00000649912.1
|
ENSG00000285723.1
|
novel protein |
| chr9_-_21368057 | 0.02 |
ENST00000449498.2
|
IFNA13
|
interferon alpha 13 |
| chr2_+_176116768 | 0.02 |
ENST00000249501.5
|
HOXD10
|
homeobox D10 |
| chr10_+_18340821 | 0.02 |
ENST00000612743.1
ENST00000612134.4 |
CACNB2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chr5_-_112419251 | 0.02 |
ENST00000261486.6
|
EPB41L4A
|
erythrocyte membrane protein band 4.1 like 4A |
| chrX_+_37990773 | 0.02 |
ENST00000341016.5
|
H2AP
|
H2A.P histone |
| chr4_-_69639642 | 0.02 |
ENST00000604629.6
ENST00000604021.1 |
UGT2A2
|
UDP glucuronosyltransferase family 2 member A2 |
| chr6_-_44313306 | 0.02 |
ENST00000244571.5
|
AARS2
|
alanyl-tRNA synthetase 2, mitochondrial |
| chr22_-_28711931 | 0.02 |
ENST00000434810.5
ENST00000456369.5 |
CHEK2
|
checkpoint kinase 2 |
| chr2_-_162838728 | 0.02 |
ENST00000328032.8
ENST00000332142.10 |
KCNH7
|
potassium voltage-gated channel subfamily H member 7 |
| chr15_-_64093746 | 0.02 |
ENST00000557835.5
ENST00000380290.7 ENST00000300030.8 ENST00000559950.1 |
CIAO2A
|
cytosolic iron-sulfur assembly component 2A |
| chr8_-_69834970 | 0.02 |
ENST00000260126.9
|
SLCO5A1
|
solute carrier organic anion transporter family member 5A1 |
| chr1_+_197268222 | 0.02 |
ENST00000367400.8
ENST00000638467.1 ENST00000367399.6 |
CRB1
|
crumbs cell polarity complex component 1 |
| chr10_+_96304392 | 0.02 |
ENST00000630152.1
|
DNTT
|
DNA nucleotidylexotransferase |
| chr13_-_23433676 | 0.02 |
ENST00000682547.1
ENST00000455470.6 ENST00000382292.9 |
SACS
|
sacsin molecular chaperone |
| chr6_+_142147162 | 0.02 |
ENST00000452973.6
ENST00000620996.4 ENST00000367621.1 ENST00000367630.9 |
VTA1
|
vesicle trafficking 1 |
| chr3_+_97821984 | 0.02 |
ENST00000389622.7
|
CRYBG3
|
crystallin beta-gamma domain containing 3 |
| chr10_+_12129637 | 0.02 |
ENST00000379051.5
ENST00000379033.7 ENST00000441368.5 ENST00000298428.14 ENST00000304267.12 ENST00000379020.8 ENST00000379017.7 |
SEC61A2
|
SEC61 translocon subunit alpha 2 |
| chr7_-_105269007 | 0.02 |
ENST00000357311.7
|
SRPK2
|
SRSF protein kinase 2 |
| chr1_-_686673 | 0.02 |
ENST00000332831.4
|
OR4F16
|
olfactory receptor family 4 subfamily F member 16 |
| chr10_-_17129786 | 0.01 |
ENST00000377833.10
|
CUBN
|
cubilin |
| chr1_+_149782671 | 0.01 |
ENST00000444948.5
ENST00000369168.5 |
FCGR1A
|
Fc fragment of IgG receptor Ia |
| chr6_+_28349907 | 0.01 |
ENST00000252211.7
ENST00000341464.9 ENST00000377255.3 |
ZKSCAN3
|
zinc finger with KRAB and SCAN domains 3 |
| chr8_-_80029826 | 0.01 |
ENST00000519386.5
|
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr13_-_23433735 | 0.01 |
ENST00000423156.2
ENST00000683210.1 ENST00000682775.1 ENST00000684497.1 ENST00000682944.1 ENST00000683489.1 ENST00000684385.1 ENST00000683680.1 |
SACS
|
sacsin molecular chaperone |
| chr7_-_77798359 | 0.01 |
ENST00000257663.4
|
TMEM60
|
transmembrane protein 60 |
| chr18_+_24460655 | 0.01 |
ENST00000426880.2
|
HRH4
|
histamine receptor H4 |
| chr7_+_77798832 | 0.01 |
ENST00000415251.6
ENST00000275575.11 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr5_-_180072086 | 0.01 |
ENST00000261947.4
|
RNF130
|
ring finger protein 130 |
| chr5_-_139198358 | 0.01 |
ENST00000394817.7
|
SIL1
|
SIL1 nucleotide exchange factor |
| chr1_-_28058087 | 0.01 |
ENST00000373864.5
|
EYA3
|
EYA transcriptional coactivator and phosphatase 3 |
| chr3_+_33114007 | 0.01 |
ENST00000320954.11
|
CRTAP
|
cartilage associated protein |
| chr2_+_233681877 | 0.01 |
ENST00000373426.4
|
UGT1A7
|
UDP glucuronosyltransferase family 1 member A7 |
| chr12_-_86256267 | 0.01 |
ENST00000620241.4
|
MGAT4C
|
MGAT4 family member C |
| chr3_+_158801926 | 0.01 |
ENST00000622669.4
ENST00000392813.8 ENST00000415822.8 ENST00000651862.1 ENST00000264266.12 |
MFSD1
|
major facilitator superfamily domain containing 1 |
| chr12_+_80099535 | 0.01 |
ENST00000646859.1
ENST00000547103.7 |
OTOGL
|
otogelin like |
| chr5_+_52787899 | 0.01 |
ENST00000274311.3
ENST00000282588.7 |
PELO
ITGA1
|
pelota mRNA surveillance and ribosome rescue factor integrin subunit alpha 1 |
| chr14_+_77708076 | 0.01 |
ENST00000238688.9
ENST00000557342.6 ENST00000557623.5 ENST00000557431.5 ENST00000556831.5 ENST00000556375.5 ENST00000553981.1 |
SLIRP
|
SRA stem-loop interacting RNA binding protein |
| chr5_+_31193739 | 0.01 |
ENST00000514738.5
|
CDH6
|
cadherin 6 |
| chr15_+_36594868 | 0.01 |
ENST00000566807.5
ENST00000643612.1 ENST00000567389.5 ENST00000562877.5 |
CDIN1
|
CDAN1 interacting nuclease 1 |
| chr5_+_70025247 | 0.01 |
ENST00000380751.9
ENST00000380750.8 ENST00000503931.5 ENST00000506542.1 |
SERF1B
|
small EDRK-rich factor 1B |
| chr4_-_99657820 | 0.01 |
ENST00000511828.2
|
C4orf54
|
chromosome 4 open reading frame 54 |
| chr7_+_123848070 | 0.01 |
ENST00000476325.5
|
HYAL4
|
hyaluronidase 4 |
| chr17_-_41047267 | 0.01 |
ENST00000542137.1
ENST00000391419.3 |
KRTAP2-1
|
keratin associated protein 2-1 |
| chr6_+_26158115 | 0.01 |
ENST00000377777.5
ENST00000289316.2 |
H2BC5
|
H2B clustered histone 5 |
| chr18_+_24460630 | 0.01 |
ENST00000256906.5
|
HRH4
|
histamine receptor H4 |
| chr7_+_77798750 | 0.01 |
ENST00000416283.6
ENST00000422959.6 ENST00000307305.12 ENST00000424760.5 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr4_-_68245683 | 0.01 |
ENST00000332644.6
|
TMPRSS11B
|
transmembrane serine protease 11B |
| chr19_-_15815664 | 0.01 |
ENST00000641419.1
|
OR10H1
|
olfactory receptor family 10 subfamily H member 1 |
| chr6_-_26250625 | 0.01 |
ENST00000618052.2
|
H3C7
|
H3 clustered histone 7 |
| chr3_+_160225409 | 0.01 |
ENST00000326474.5
|
C3orf80
|
chromosome 3 open reading frame 80 |
| chr12_-_56741535 | 0.01 |
ENST00000647707.1
|
ENSG00000285625.1
|
novel protein |
| chr11_+_66011994 | 0.01 |
ENST00000312134.3
|
CST6
|
cystatin E/M |
| chr14_+_99481395 | 0.00 |
ENST00000389879.9
ENST00000557441.5 ENST00000555049.5 ENST00000555842.1 |
CCNK
|
cyclin K |
| chr11_-_11353241 | 0.00 |
ENST00000528848.3
|
CSNK2A3
|
casein kinase 2 alpha 3 |
| chr8_+_75539893 | 0.00 |
ENST00000674002.1
|
HNF4G
|
hepatocyte nuclear factor 4 gamma |
| chr10_+_18340699 | 0.00 |
ENST00000377329.10
|
CACNB2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chr1_-_17045219 | 0.00 |
ENST00000491274.5
|
SDHB
|
succinate dehydrogenase complex iron sulfur subunit B |
| chr5_-_177780633 | 0.00 |
ENST00000513554.5
ENST00000440605.7 |
FAM153A
|
family with sequence similarity 153 member A |
| chr20_+_43507668 | 0.00 |
ENST00000434666.5
ENST00000649084.1 ENST00000418998.7 ENST00000427442.8 ENST00000649917.1 |
L3MBTL1
|
L3MBTL histone methyl-lysine binding protein 1 |
| chr16_-_2329687 | 0.00 |
ENST00000567910.1
|
ABCA3
|
ATP binding cassette subfamily A member 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXD4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1904397 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 0.1 | 0.2 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 | 0.8 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.6 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.0 | 0.2 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.1 | GO:0010607 | negative regulation of cytoplasmic mRNA processing body assembly(GO:0010607) |
| 0.0 | 0.2 | GO:0051958 | methotrexate transport(GO:0051958) |
| 0.0 | 0.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.0 | 0.4 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 0.1 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.1 | GO:0044771 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.1 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.0 | 0.0 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.1 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.2 | GO:0051918 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) negative regulation of fibrinolysis(GO:0051918) |
| 0.0 | 0.0 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.0 | 0.2 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.1 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.0 | 0.1 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.0 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.0 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.0 | 0.0 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.0 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.2 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.0 | 0.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 1.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.3 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.0 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.1 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.0 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0052828 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.1 | 0.7 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.2 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.1 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.0 | 0.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.2 | GO:0015350 | methotrexate transporter activity(GO:0015350) |
| 0.0 | 0.2 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.0 | 0.1 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.0 | 0.0 | GO:0008988 | rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) |
| 0.0 | 0.5 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.0 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.0 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |