Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
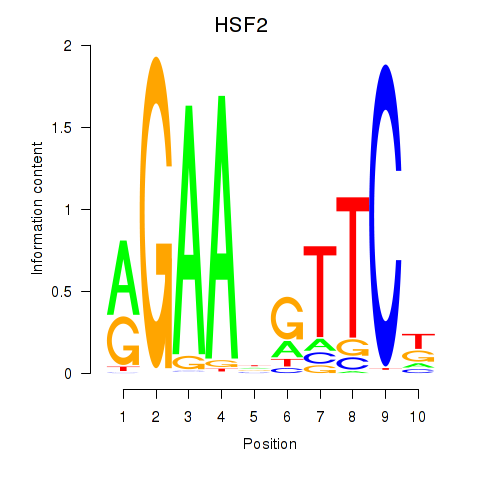
Results for HSF2
Z-value: 0.09
Transcription factors associated with HSF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HSF2
|
ENSG00000025156.13 | heat shock transcription factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HSF2 | hg38_v1_chr6_+_122399536_122399568, hg38_v1_chr6_+_122399621_122399692 | -0.37 | 3.7e-01 | Click! |
Activity profile of HSF2 motif
Sorted Z-values of HSF2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_81324130 | 0.23 |
ENST00000302824.7
|
STARD5
|
StAR related lipid transfer domain containing 5 |
| chr11_-_111910790 | 0.20 |
ENST00000533280.6
|
CRYAB
|
crystallin alpha B |
| chr11_-_111910830 | 0.20 |
ENST00000526167.5
ENST00000651650.1 |
CRYAB
|
crystallin alpha B |
| chr11_-_111910888 | 0.19 |
ENST00000525823.1
ENST00000528961.6 |
CRYAB
|
crystallin alpha B |
| chr3_+_45026296 | 0.19 |
ENST00000296130.5
|
CLEC3B
|
C-type lectin domain family 3 member B |
| chr16_-_9943182 | 0.18 |
ENST00000535259.6
|
GRIN2A
|
glutamate ionotropic receptor NMDA type subunit 2A |
| chr4_-_185956348 | 0.13 |
ENST00000431902.5
ENST00000284776.11 ENST00000415274.5 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr1_-_168729187 | 0.12 |
ENST00000367817.4
|
DPT
|
dermatopontin |
| chr8_-_22693469 | 0.11 |
ENST00000317216.3
|
EGR3
|
early growth response 3 |
| chr4_-_185956652 | 0.11 |
ENST00000355634.9
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr7_+_130486171 | 0.10 |
ENST00000341441.9
ENST00000416162.7 |
MEST
|
mesoderm specific transcript |
| chr7_+_130486324 | 0.10 |
ENST00000427521.6
ENST00000378576.9 |
MEST
|
mesoderm specific transcript |
| chr6_-_109094819 | 0.09 |
ENST00000436639.6
|
SESN1
|
sestrin 1 |
| chr14_-_52069039 | 0.09 |
ENST00000216286.10
|
NID2
|
nidogen 2 |
| chr14_-_52069228 | 0.08 |
ENST00000617139.4
|
NID2
|
nidogen 2 |
| chr7_-_80919017 | 0.08 |
ENST00000265361.8
|
SEMA3C
|
semaphorin 3C |
| chr8_+_133113483 | 0.08 |
ENST00000521107.1
|
TG
|
thyroglobulin |
| chr8_-_119592954 | 0.08 |
ENST00000522167.5
|
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr19_-_18538371 | 0.07 |
ENST00000596015.1
|
FKBP8
|
FKBP prolyl isomerase 8 |
| chr3_-_112641128 | 0.07 |
ENST00000206423.8
|
CCDC80
|
coiled-coil domain containing 80 |
| chr16_+_176659 | 0.07 |
ENST00000320868.9
ENST00000397797.1 |
HBA1
|
hemoglobin subunit alpha 1 |
| chr11_+_46295126 | 0.07 |
ENST00000534787.1
|
CREB3L1
|
cAMP responsive element binding protein 3 like 1 |
| chr19_-_55180010 | 0.06 |
ENST00000589172.5
|
SYT5
|
synaptotagmin 5 |
| chrX_+_136148440 | 0.06 |
ENST00000627383.2
ENST00000630084.2 |
FHL1
|
four and a half LIM domains 1 |
| chrX_+_136197020 | 0.06 |
ENST00000370676.7
|
FHL1
|
four and a half LIM domains 1 |
| chr13_+_45702306 | 0.06 |
ENST00000533564.1
ENST00000310521.6 |
CBY2
|
chibby family member 2 |
| chr13_+_45702411 | 0.05 |
ENST00000610924.1
|
CBY2
|
chibby family member 2 |
| chr16_+_55566668 | 0.05 |
ENST00000457326.3
|
CAPNS2
|
calpain small subunit 2 |
| chr1_+_18631513 | 0.05 |
ENST00000400661.3
|
PAX7
|
paired box 7 |
| chr6_+_29396555 | 0.05 |
ENST00000623183.1
|
OR12D2
|
olfactory receptor family 12 subfamily D member 2 |
| chr7_+_76302665 | 0.05 |
ENST00000248553.7
ENST00000674638.1 ENST00000674547.1 ENST00000675226.1 ENST00000675538.1 ENST00000676231.1 ENST00000675134.1 ENST00000675906.1 ENST00000674650.1 |
HSPB1
|
heat shock protein family B (small) member 1 |
| chrX_+_136196750 | 0.05 |
ENST00000539015.5
|
FHL1
|
four and a half LIM domains 1 |
| chrX_+_136197039 | 0.05 |
ENST00000370683.6
|
FHL1
|
four and a half LIM domains 1 |
| chr4_+_76011171 | 0.05 |
ENST00000513353.5
ENST00000341029.9 |
ART3
|
ADP-ribosyltransferase 3 (inactive) |
| chr7_-_14841267 | 0.04 |
ENST00000406247.7
ENST00000399322.7 |
DGKB
|
diacylglycerol kinase beta |
| chrX_+_106920393 | 0.04 |
ENST00000336803.2
|
CLDN2
|
claudin 2 |
| chr3_-_112641292 | 0.04 |
ENST00000439685.6
|
CCDC80
|
coiled-coil domain containing 80 |
| chr3_+_159069252 | 0.04 |
ENST00000640015.1
ENST00000476809.7 ENST00000485419.7 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr11_+_66509079 | 0.04 |
ENST00000419755.3
|
ENSG00000256349.1
|
novel protein |
| chr11_-_83286377 | 0.04 |
ENST00000455220.6
ENST00000529689.5 |
CCDC90B
|
coiled-coil domain containing 90B |
| chr1_-_120176450 | 0.04 |
ENST00000578049.4
|
SEC22B
|
SEC22 homolog B, vesicle trafficking protein |
| chr11_-_83285965 | 0.04 |
ENST00000529073.5
ENST00000529611.5 |
CCDC90B
|
coiled-coil domain containing 90B |
| chr1_-_184037695 | 0.04 |
ENST00000361927.9
ENST00000649786.1 |
COLGALT2
|
collagen beta(1-O)galactosyltransferase 2 |
| chr11_+_1834415 | 0.04 |
ENST00000381968.7
ENST00000381978.7 |
SYT8
|
synaptotagmin 8 |
| chr11_-_83286328 | 0.03 |
ENST00000525503.5
|
CCDC90B
|
coiled-coil domain containing 90B |
| chr5_+_140125935 | 0.03 |
ENST00000333305.5
|
IGIP
|
IgA inducing protein |
| chr8_-_26513865 | 0.03 |
ENST00000522362.7
|
PNMA2
|
PNMA family member 2 |
| chr11_+_1834804 | 0.03 |
ENST00000341958.3
|
SYT8
|
synaptotagmin 8 |
| chr11_+_86302211 | 0.03 |
ENST00000533986.5
ENST00000278483.8 |
HIKESHI
|
heat shock protein nuclear import factor hikeshi |
| chr10_+_119726118 | 0.03 |
ENST00000369081.3
ENST00000648262.1 |
INPP5F
|
inositol polyphosphate-5-phosphatase F |
| chr19_+_9250930 | 0.03 |
ENST00000456448.3
|
OR7E24
|
olfactory receptor family 7 subfamily E member 24 |
| chr7_+_31529675 | 0.03 |
ENST00000451887.6
ENST00000454513.5 |
ITPRID1
|
ITPR interacting domain containing 1 |
| chr1_-_205994439 | 0.03 |
ENST00000617991.4
|
RAB7B
|
RAB7B, member RAS oncogene family |
| chr3_-_57079287 | 0.03 |
ENST00000338458.8
ENST00000468727.5 |
ARHGEF3
|
Rho guanine nucleotide exchange factor 3 |
| chr1_-_119648165 | 0.03 |
ENST00000421812.3
|
ZNF697
|
zinc finger protein 697 |
| chr4_+_182448995 | 0.03 |
ENST00000510504.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr16_-_30445865 | 0.03 |
ENST00000478753.5
|
SEPHS2
|
selenophosphate synthetase 2 |
| chrX_-_107982370 | 0.03 |
ENST00000302917.1
|
TEX13B
|
testis expressed 13B |
| chr3_-_129688691 | 0.03 |
ENST00000432054.6
|
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chr3_+_96814552 | 0.02 |
ENST00000470610.6
ENST00000389672.9 |
EPHA6
|
EPH receptor A6 |
| chr9_-_4666495 | 0.02 |
ENST00000475086.5
|
SPATA6L
|
spermatogenesis associated 6 like |
| chr12_+_119178920 | 0.02 |
ENST00000281938.7
|
HSPB8
|
heat shock protein family B (small) member 8 |
| chr17_-_1628341 | 0.02 |
ENST00000571650.5
|
SLC43A2
|
solute carrier family 43 member 2 |
| chr12_+_119178953 | 0.02 |
ENST00000674542.1
|
HSPB8
|
heat shock protein family B (small) member 8 |
| chr5_+_57173948 | 0.02 |
ENST00000424459.7
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr11_-_65557740 | 0.02 |
ENST00000526927.5
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr17_-_76141240 | 0.02 |
ENST00000322957.7
|
FOXJ1
|
forkhead box J1 |
| chr3_-_184017863 | 0.02 |
ENST00000427120.6
ENST00000334444.11 ENST00000392579.6 ENST00000382494.6 ENST00000265586.10 ENST00000446941.2 |
ABCC5
|
ATP binding cassette subfamily C member 5 |
| chr9_+_34989641 | 0.02 |
ENST00000453597.8
ENST00000312316.9 ENST00000458263.6 ENST00000537321.5 ENST00000682809.1 ENST00000684748.1 |
DNAJB5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr2_-_174682854 | 0.02 |
ENST00000409415.7
ENST00000359761.7 ENST00000272746.9 |
WIPF1
|
WAS/WASL interacting protein family member 1 |
| chr1_+_192575765 | 0.02 |
ENST00000469578.2
ENST00000367459.8 |
RGS1
|
regulator of G protein signaling 1 |
| chr2_-_177263522 | 0.02 |
ENST00000448782.5
ENST00000446151.6 |
NFE2L2
|
nuclear factor, erythroid 2 like 2 |
| chr9_+_5629025 | 0.02 |
ENST00000251879.10
ENST00000414202.7 ENST00000418622.7 |
RIC1
|
RIC1 homolog, RAB6A GEF complex partner 1 |
| chr10_+_102776237 | 0.02 |
ENST00000369889.5
|
WBP1L
|
WW domain binding protein 1 like |
| chr2_+_218959635 | 0.02 |
ENST00000302625.6
|
CDK5R2
|
cyclin dependent kinase 5 regulatory subunit 2 |
| chr14_-_25010604 | 0.02 |
ENST00000550887.5
|
STXBP6
|
syntaxin binding protein 6 |
| chr5_+_57174016 | 0.02 |
ENST00000514387.6
ENST00000506184.7 |
GPBP1
|
GC-rich promoter binding protein 1 |
| chr18_+_46917561 | 0.02 |
ENST00000683218.1
|
KATNAL2
|
katanin catalytic subunit A1 like 2 |
| chr11_+_72080803 | 0.02 |
ENST00000423494.6
ENST00000539587.6 ENST00000536917.2 ENST00000538478.5 ENST00000324866.11 ENST00000643715.1 ENST00000439209.5 |
LRTOMT
ENSG00000284922.2
|
leucine rich transmembrane and O-methyltransferase domain containing leucine rich transmembrane and O-methyltransferase domain containing |
| chr5_+_119356011 | 0.02 |
ENST00000504771.3
ENST00000415806.2 |
TNFAIP8
|
TNF alpha induced protein 8 |
| chr11_+_59511539 | 0.02 |
ENST00000641962.1
|
OR4D9
|
olfactory receptor family 4 subfamily D member 9 |
| chr2_+_11542662 | 0.02 |
ENST00000389825.7
ENST00000381483.6 |
GREB1
|
growth regulating estrogen receptor binding 1 |
| chrX_+_15749848 | 0.02 |
ENST00000479740.5
ENST00000454127.2 |
CA5B
|
carbonic anhydrase 5B |
| chr11_+_72080595 | 0.02 |
ENST00000647530.1
ENST00000539271.6 ENST00000642510.1 |
LRTOMT
|
leucine rich transmembrane and O-methyltransferase domain containing |
| chr20_+_58852710 | 0.02 |
ENST00000676826.2
ENST00000371100.9 |
GNAS
|
GNAS complex locus |
| chr19_+_14031746 | 0.02 |
ENST00000263379.4
|
IL27RA
|
interleukin 27 receptor subunit alpha |
| chr17_-_78577383 | 0.02 |
ENST00000389840.7
|
DNAH17
|
dynein axonemal heavy chain 17 |
| chr9_-_34589716 | 0.02 |
ENST00000378980.8
|
CNTFR
|
ciliary neurotrophic factor receptor |
| chr17_-_41505597 | 0.02 |
ENST00000336861.7
ENST00000246635.8 ENST00000587544.5 ENST00000587435.1 |
KRT13
|
keratin 13 |
| chr12_-_44875647 | 0.02 |
ENST00000395487.6
|
NELL2
|
neural EGFL like 2 |
| chr14_-_77423517 | 0.02 |
ENST00000555603.1
|
NOXRED1
|
NADP dependent oxidoreductase domain containing 1 |
| chr17_-_58417521 | 0.02 |
ENST00000584437.5
ENST00000407977.7 |
RNF43
|
ring finger protein 43 |
| chr9_-_34589701 | 0.01 |
ENST00000351266.8
|
CNTFR
|
ciliary neurotrophic factor receptor |
| chr12_+_57745017 | 0.01 |
ENST00000547992.5
ENST00000552816.5 ENST00000257910.8 ENST00000547472.5 |
TSPAN31
|
tetraspanin 31 |
| chr17_-_58417547 | 0.01 |
ENST00000577716.5
|
RNF43
|
ring finger protein 43 |
| chr12_-_44876294 | 0.01 |
ENST00000429094.7
ENST00000551601.5 ENST00000549027.5 ENST00000452445.6 |
NELL2
|
neural EGFL like 2 |
| chr6_+_32741382 | 0.01 |
ENST00000374940.4
|
HLA-DQA2
|
major histocompatibility complex, class II, DQ alpha 2 |
| chr12_+_52010485 | 0.01 |
ENST00000552049.5
ENST00000546756.1 |
TAMALIN
|
trafficking regulator and scaffold protein tamalin |
| chr9_+_123033660 | 0.01 |
ENST00000616002.3
|
GPR21
|
G protein-coupled receptor 21 |
| chr2_+_28392802 | 0.01 |
ENST00000379619.5
ENST00000264716.9 |
FOSL2
|
FOS like 2, AP-1 transcription factor subunit |
| chr12_-_122526929 | 0.01 |
ENST00000331738.12
ENST00000528279.1 ENST00000344591.8 ENST00000526560.6 |
RSRC2
|
arginine and serine rich coiled-coil 2 |
| chr18_+_3451585 | 0.01 |
ENST00000551541.5
|
TGIF1
|
TGFB induced factor homeobox 1 |
| chr17_-_7394514 | 0.01 |
ENST00000571802.1
ENST00000619711.5 ENST00000576201.5 ENST00000573213.1 ENST00000324822.15 |
PLSCR3
|
phospholipid scramblase 3 |
| chr6_+_106087580 | 0.01 |
ENST00000424894.1
ENST00000648754.1 |
PRDM1
|
PR/SET domain 1 |
| chr4_-_48134200 | 0.01 |
ENST00000264316.9
|
TXK
|
TXK tyrosine kinase |
| chr12_+_106684696 | 0.01 |
ENST00000229387.6
|
RFX4
|
regulatory factor X4 |
| chr12_+_6535278 | 0.01 |
ENST00000396858.5
|
GAPDH
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr11_-_73142308 | 0.01 |
ENST00000409418.9
|
FCHSD2
|
FCH and double SH3 domains 2 |
| chr3_+_172040554 | 0.01 |
ENST00000336824.8
ENST00000423424.5 |
FNDC3B
|
fibronectin type III domain containing 3B |
| chr18_+_3451647 | 0.01 |
ENST00000345133.9
ENST00000330513.10 ENST00000549546.5 |
TGIF1
|
TGFB induced factor homeobox 1 |
| chr11_+_59511368 | 0.01 |
ENST00000641278.1
|
OR4D9
|
olfactory receptor family 4 subfamily D member 9 |
| chr11_+_66002754 | 0.01 |
ENST00000527348.1
|
BANF1
|
BAF nuclear assembly factor 1 |
| chr3_-_48595267 | 0.01 |
ENST00000328333.12
ENST00000681320.1 |
COL7A1
|
collagen type VII alpha 1 chain |
| chr11_-_124310837 | 0.01 |
ENST00000357821.2
|
OR8D1
|
olfactory receptor family 8 subfamily D member 1 |
| chr7_-_150955796 | 0.01 |
ENST00000330883.9
|
KCNH2
|
potassium voltage-gated channel subfamily H member 2 |
| chr11_+_66002475 | 0.01 |
ENST00000312175.7
ENST00000533166.5 |
BANF1
|
BAF nuclear assembly factor 1 |
| chr6_+_146543824 | 0.01 |
ENST00000367495.4
|
RAB32
|
RAB32, member RAS oncogene family |
| chr2_+_197500398 | 0.01 |
ENST00000604458.1
|
HSPE1-MOB4
|
HSPE1-MOB4 readthrough |
| chr19_+_17305801 | 0.01 |
ENST00000602206.1
ENST00000252602.2 |
MRPL34
|
mitochondrial ribosomal protein L34 |
| chr9_-_95317671 | 0.01 |
ENST00000490972.7
ENST00000647778.1 ENST00000649611.1 ENST00000289081.8 |
FANCC
|
FA complementation group C |
| chr6_-_53148822 | 0.01 |
ENST00000259803.8
|
GCM1
|
glial cells missing transcription factor 1 |
| chr10_-_1200360 | 0.01 |
ENST00000381310.7
ENST00000381305.1 |
ADARB2
|
adenosine deaminase RNA specific B2 (inactive) |
| chr1_-_101996919 | 0.01 |
ENST00000370103.9
|
OLFM3
|
olfactomedin 3 |
| chr7_+_38977904 | 0.01 |
ENST00000518318.7
ENST00000403058.6 |
POU6F2
|
POU class 6 homeobox 2 |
| chr20_+_33168148 | 0.01 |
ENST00000354932.6
|
BPIFA2
|
BPI fold containing family A member 2 |
| chr2_-_96740034 | 0.01 |
ENST00000264963.9
ENST00000377079.8 |
LMAN2L
|
lectin, mannose binding 2 like |
| chr7_-_83649097 | 0.01 |
ENST00000643230.2
|
SEMA3E
|
semaphorin 3E |
| chr2_-_74343397 | 0.01 |
ENST00000394019.6
ENST00000377634.8 ENST00000436454.1 |
SLC4A5
|
solute carrier family 4 member 5 |
| chrX_+_51743395 | 0.01 |
ENST00000340438.6
|
GSPT2
|
G1 to S phase transition 2 |
| chr1_-_111989155 | 0.01 |
ENST00000315987.6
|
KCND3
|
potassium voltage-gated channel subfamily D member 3 |
| chr2_-_216695540 | 0.01 |
ENST00000233813.5
|
IGFBP5
|
insulin like growth factor binding protein 5 |
| chr18_+_24139013 | 0.01 |
ENST00000399481.6
ENST00000327201.10 |
CABYR
|
calcium binding tyrosine phosphorylation regulated |
| chr10_-_24721866 | 0.01 |
ENST00000416305.1
ENST00000320481.10 |
ARHGAP21
|
Rho GTPase activating protein 21 |
| chr7_-_78771265 | 0.01 |
ENST00000630991.2
ENST00000629359.2 |
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr15_-_43493076 | 0.01 |
ENST00000413546.1
|
TP53BP1
|
tumor protein p53 binding protein 1 |
| chr9_-_89178810 | 0.01 |
ENST00000375835.9
|
SHC3
|
SHC adaptor protein 3 |
| chr1_-_36450410 | 0.01 |
ENST00000356637.9
ENST00000354267.3 ENST00000235532.9 |
OSCP1
|
organic solute carrier partner 1 |
| chr6_-_31815244 | 0.01 |
ENST00000375654.5
|
HSPA1L
|
heat shock protein family A (Hsp70) member 1 like |
| chr14_+_103334339 | 0.01 |
ENST00000558316.5
ENST00000558265.5 |
EIF5
|
eukaryotic translation initiation factor 5 |
| chr15_+_67543178 | 0.01 |
ENST00000395476.6
|
MAP2K5
|
mitogen-activated protein kinase kinase 5 |
| chr19_-_48746797 | 0.01 |
ENST00000602105.1
ENST00000332955.7 |
IZUMO1
|
izumo sperm-egg fusion 1 |
| chr9_-_4666347 | 0.01 |
ENST00000381890.9
ENST00000682582.1 |
SPATA6L
|
spermatogenesis associated 6 like |
| chr6_-_32154326 | 0.01 |
ENST00000475826.1
ENST00000485392.5 ENST00000494332.5 ENST00000498575.1 ENST00000428778.5 |
ENSG00000284954.1
ENSG00000285085.1
|
novel transcript novel protein |
| chr5_+_80035341 | 0.01 |
ENST00000350881.6
|
THBS4
|
thrombospondin 4 |
| chr7_+_130344810 | 0.01 |
ENST00000497503.5
ENST00000463587.5 ENST00000461828.5 ENST00000474905.6 ENST00000494311.1 ENST00000466363.6 |
CPA5
|
carboxypeptidase A5 |
| chr7_+_157336988 | 0.01 |
ENST00000262177.9
ENST00000417758.5 ENST00000443280.5 |
DNAJB6
|
DnaJ heat shock protein family (Hsp40) member B6 |
| chr1_-_197775435 | 0.01 |
ENST00000620048.6
|
DENND1B
|
DENN domain containing 1B |
| chr16_+_55655960 | 0.01 |
ENST00000568943.6
|
SLC6A2
|
solute carrier family 6 member 2 |
| chr11_-_133956957 | 0.01 |
ENST00000533871.8
|
IGSF9B
|
immunoglobulin superfamily member 9B |
| chr9_+_125747345 | 0.01 |
ENST00000342287.9
ENST00000373489.10 ENST00000373487.8 |
PBX3
|
PBX homeobox 3 |
| chr21_+_43728851 | 0.01 |
ENST00000327574.4
|
PDXK
|
pyridoxal kinase |
| chr7_-_78771108 | 0.00 |
ENST00000626691.2
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr4_+_122239965 | 0.00 |
ENST00000446180.5
|
KIAA1109
|
KIAA1109 |
| chr8_+_86866415 | 0.00 |
ENST00000518476.6
|
CNBD1
|
cyclic nucleotide binding domain containing 1 |
| chr7_+_130344837 | 0.00 |
ENST00000485477.5
ENST00000431780.6 |
CPA5
|
carboxypeptidase A5 |
| chr14_+_37622065 | 0.00 |
ENST00000553443.5
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chr9_-_33025088 | 0.00 |
ENST00000436040.7
|
APTX
|
aprataxin |
| chr11_+_66002225 | 0.00 |
ENST00000445560.6
ENST00000530204.1 |
BANF1
|
BAF nuclear assembly factor 1 |
| chr2_+_161416273 | 0.00 |
ENST00000389554.8
|
TBR1
|
T-box brain transcription factor 1 |
| chr10_+_120457197 | 0.00 |
ENST00000398250.6
|
PLPP4
|
phospholipid phosphatase 4 |
| chr19_-_344786 | 0.00 |
ENST00000264819.7
|
MIER2
|
MIER family member 2 |
| chr3_-_47513677 | 0.00 |
ENST00000296149.9
|
ELP6
|
elongator acetyltransferase complex subunit 6 |
| chr8_-_80171106 | 0.00 |
ENST00000519303.6
|
TPD52
|
tumor protein D52 |
| chr1_-_36450279 | 0.00 |
ENST00000445843.7
|
OSCP1
|
organic solute carrier partner 1 |
| chr13_+_111115303 | 0.00 |
ENST00000646102.2
ENST00000449979.5 |
ARHGEF7
|
Rho guanine nucleotide exchange factor 7 |
| chr20_+_18813777 | 0.00 |
ENST00000377428.4
|
SCP2D1
|
SCP2 sterol binding domain containing 1 |
| chr9_-_33025052 | 0.00 |
ENST00000673248.1
|
APTX
|
aprataxin |
| chr6_-_123636923 | 0.00 |
ENST00000334268.9
|
TRDN
|
triadin |
| chr4_-_76421121 | 0.00 |
ENST00000682701.1
|
CCDC158
|
coiled-coil domain containing 158 |
| chr19_-_55180242 | 0.00 |
ENST00000592470.1
ENST00000354308.8 |
SYT5
|
synaptotagmin 5 |
| chr5_-_160312524 | 0.00 |
ENST00000520748.1
ENST00000257536.13 ENST00000393977.7 |
CCNJL
|
cyclin J like |
| chr15_+_67542668 | 0.00 |
ENST00000178640.10
|
MAP2K5
|
mitogen-activated protein kinase kinase 5 |
| chr10_-_103153609 | 0.00 |
ENST00000675985.1
|
NT5C2
|
5'-nucleotidase, cytosolic II |
| chr22_+_40857076 | 0.00 |
ENST00000614001.1
ENST00000357137.9 |
XPNPEP3
|
X-prolyl aminopeptidase 3 |
| chr18_-_812230 | 0.00 |
ENST00000314574.5
|
YES1
|
YES proto-oncogene 1, Src family tyrosine kinase |
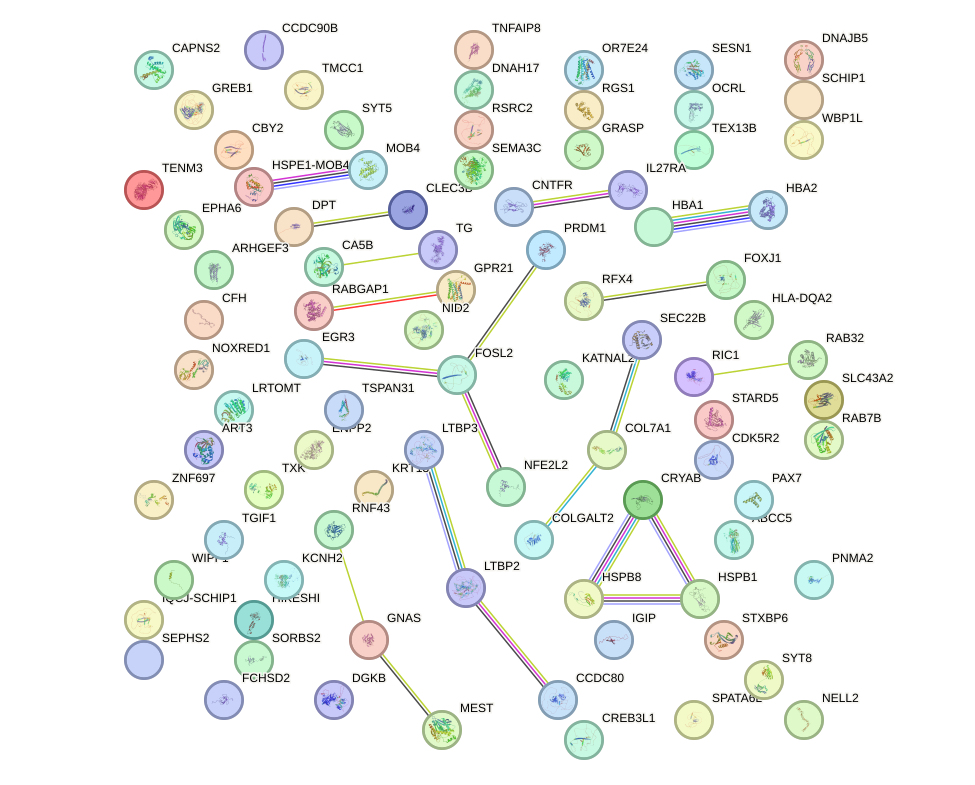
Network of associatons between targets according to the STRING database.
First level regulatory network of HSF2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.0 | 0.6 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0014813 | skeletal muscle satellite cell commitment(GO:0014813) |
| 0.0 | 0.2 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.1 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.0 | 0.1 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.2 | GO:0035376 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.0 | 0.1 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.2 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.2 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.1 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |