Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
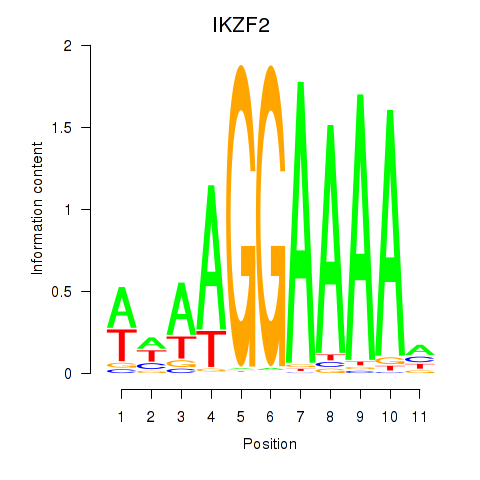
Results for IKZF2
Z-value: 1.24
Transcription factors associated with IKZF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IKZF2
|
ENSG00000030419.17 | IKAROS family zinc finger 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IKZF2 | hg38_v1_chr2_-_213151590_213151619 | 0.82 | 1.3e-02 | Click! |
Activity profile of IKZF2 motif
Sorted Z-values of IKZF2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_188974364 | 2.68 |
ENST00000304636.9
ENST00000317840.9 |
COL3A1
|
collagen type III alpha 1 chain |
| chr1_-_56579555 | 2.26 |
ENST00000371250.4
|
PLPP3
|
phospholipid phosphatase 3 |
| chr12_-_91179472 | 2.15 |
ENST00000550099.5
ENST00000546391.5 |
DCN
|
decorin |
| chr12_-_91179355 | 1.83 |
ENST00000550563.5
ENST00000546370.5 |
DCN
|
decorin |
| chr8_+_96584920 | 1.74 |
ENST00000521590.5
|
SDC2
|
syndecan 2 |
| chr7_+_94394886 | 1.57 |
ENST00000297268.11
ENST00000620463.1 |
COL1A2
|
collagen type I alpha 2 chain |
| chr3_+_12351493 | 1.30 |
ENST00000683699.1
|
PPARG
|
peroxisome proliferator activated receptor gamma |
| chr4_+_123399488 | 1.30 |
ENST00000394339.2
|
SPRY1
|
sprouty RTK signaling antagonist 1 |
| chr17_-_69141878 | 1.20 |
ENST00000590645.1
ENST00000284425.7 |
ABCA6
|
ATP binding cassette subfamily A member 6 |
| chr13_-_37598750 | 1.20 |
ENST00000379743.8
ENST00000379742.4 ENST00000379749.8 ENST00000379747.9 ENST00000541179.5 ENST00000541481.5 |
POSTN
|
periostin |
| chr3_+_12351470 | 1.16 |
ENST00000287820.10
|
PPARG
|
peroxisome proliferator activated receptor gamma |
| chr12_-_55688891 | 1.12 |
ENST00000557555.2
|
ITGA7
|
integrin subunit alpha 7 |
| chr5_+_32711313 | 1.02 |
ENST00000265074.13
|
NPR3
|
natriuretic peptide receptor 3 |
| chr5_+_32710630 | 0.99 |
ENST00000326958.5
|
NPR3
|
natriuretic peptide receptor 3 |
| chr7_+_93921720 | 0.91 |
ENST00000248564.6
|
GNG11
|
G protein subunit gamma 11 |
| chr2_+_209580024 | 0.90 |
ENST00000392194.5
|
MAP2
|
microtubule associated protein 2 |
| chr8_-_107498041 | 0.78 |
ENST00000297450.7
|
ANGPT1
|
angiopoietin 1 |
| chr14_+_51860391 | 0.72 |
ENST00000335281.8
|
GNG2
|
G protein subunit gamma 2 |
| chr2_+_209579399 | 0.71 |
ENST00000360351.8
|
MAP2
|
microtubule associated protein 2 |
| chr2_+_209579598 | 0.70 |
ENST00000445941.5
ENST00000673860.1 |
MAP2
|
microtubule associated protein 2 |
| chr2_-_189179754 | 0.67 |
ENST00000374866.9
ENST00000618828.1 |
COL5A2
|
collagen type V alpha 2 chain |
| chrX_+_81202066 | 0.65 |
ENST00000373212.6
|
SH3BGRL
|
SH3 domain binding glutamate rich protein like |
| chr11_-_1757452 | 0.64 |
ENST00000427721.3
|
ENSG00000250644.3
|
novel protein |
| chr3_-_112610262 | 0.62 |
ENST00000479368.1
|
CCDC80
|
coiled-coil domain containing 80 |
| chr9_-_70869076 | 0.61 |
ENST00000677594.1
|
TRPM3
|
transient receptor potential cation channel subfamily M member 3 |
| chr1_+_78649818 | 0.61 |
ENST00000370747.9
ENST00000438486.1 |
IFI44
|
interferon induced protein 44 |
| chr15_-_82045998 | 0.57 |
ENST00000329713.5
|
MEX3B
|
mex-3 RNA binding family member B |
| chr5_-_111756245 | 0.57 |
ENST00000447165.6
|
NREP
|
neuronal regeneration related protein |
| chr6_+_31655888 | 0.56 |
ENST00000375916.4
|
APOM
|
apolipoprotein M |
| chr15_-_82046119 | 0.55 |
ENST00000558133.1
|
MEX3B
|
mex-3 RNA binding family member B |
| chr3_-_18424533 | 0.55 |
ENST00000417717.6
|
SATB1
|
SATB homeobox 1 |
| chr3_+_69763726 | 0.52 |
ENST00000448226.9
|
MITF
|
melanocyte inducing transcription factor |
| chr4_-_185775271 | 0.52 |
ENST00000430503.5
ENST00000319454.10 ENST00000450341.5 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chrX_-_136780925 | 0.52 |
ENST00000250617.7
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor 6 |
| chr2_-_101151253 | 0.51 |
ENST00000376840.8
ENST00000409318.2 |
TBC1D8
|
TBC1 domain family member 8 |
| chrX_+_103776493 | 0.49 |
ENST00000433491.5
ENST00000612423.4 ENST00000443502.5 |
PLP1
|
proteolipid protein 1 |
| chr4_+_73481737 | 0.48 |
ENST00000226355.5
|
AFM
|
afamin |
| chrX_+_103776831 | 0.48 |
ENST00000621218.5
ENST00000619236.1 |
PLP1
|
proteolipid protein 1 |
| chr4_-_156970903 | 0.48 |
ENST00000422544.2
|
PDGFC
|
platelet derived growth factor C |
| chr3_-_18438767 | 0.46 |
ENST00000454909.6
|
SATB1
|
SATB homeobox 1 |
| chr3_+_46577778 | 0.46 |
ENST00000296145.6
|
TDGF1
|
teratocarcinoma-derived growth factor 1 |
| chr11_+_126283059 | 0.46 |
ENST00000392679.6
ENST00000392678.7 ENST00000392680.6 |
TIRAP
|
TIR domain containing adaptor protein |
| chr2_+_157257687 | 0.46 |
ENST00000259056.5
|
GALNT5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr2_-_118847638 | 0.46 |
ENST00000295206.7
|
EN1
|
engrailed homeobox 1 |
| chr2_+_209579429 | 0.45 |
ENST00000361559.8
|
MAP2
|
microtubule associated protein 2 |
| chr12_+_65279445 | 0.44 |
ENST00000642404.1
|
MSRB3
|
methionine sulfoxide reductase B3 |
| chrX_+_55452119 | 0.44 |
ENST00000342972.3
|
MAGEH1
|
MAGE family member H1 |
| chr1_+_60865259 | 0.44 |
ENST00000371191.5
|
NFIA
|
nuclear factor I A |
| chr8_-_71356653 | 0.43 |
ENST00000388742.8
ENST00000388740.4 |
EYA1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr2_-_175005357 | 0.42 |
ENST00000409156.7
ENST00000444573.2 ENST00000409900.9 |
CHN1
|
chimerin 1 |
| chr8_-_71356511 | 0.42 |
ENST00000419131.6
ENST00000388743.6 |
EYA1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr2_+_27078598 | 0.42 |
ENST00000380320.9
|
EMILIN1
|
elastin microfibril interfacer 1 |
| chr12_+_8157034 | 0.42 |
ENST00000396570.7
|
ZNF705A
|
zinc finger protein 705A |
| chr15_+_76336755 | 0.41 |
ENST00000290759.9
|
ISL2
|
ISL LIM homeobox 2 |
| chr12_-_10453330 | 0.41 |
ENST00000347831.9
ENST00000359151.8 |
KLRC1
|
killer cell lectin like receptor C1 |
| chr4_-_185812209 | 0.39 |
ENST00000393523.6
ENST00000393528.7 ENST00000449407.6 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr5_+_173889337 | 0.39 |
ENST00000520867.5
ENST00000334035.9 |
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr19_+_49930219 | 0.39 |
ENST00000596658.1
|
ATF5
|
activating transcription factor 5 |
| chr6_-_15586006 | 0.39 |
ENST00000462989.6
|
DTNBP1
|
dystrobrevin binding protein 1 |
| chr9_-_70869029 | 0.39 |
ENST00000361823.9
ENST00000377101.5 ENST00000360823.6 ENST00000377105.5 |
TRPM3
|
transient receptor potential cation channel subfamily M member 3 |
| chr1_-_153985366 | 0.38 |
ENST00000614713.4
|
RAB13
|
RAB13, member RAS oncogene family |
| chr10_+_97319250 | 0.36 |
ENST00000371021.5
|
FRAT1
|
FRAT regulator of WNT signaling pathway 1 |
| chr2_+_102070360 | 0.35 |
ENST00000409929.5
ENST00000424272.5 |
IL1R1
|
interleukin 1 receptor type 1 |
| chr8_+_76681208 | 0.35 |
ENST00000651372.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr19_-_43205551 | 0.35 |
ENST00000599391.1
ENST00000244295.13 ENST00000596907.5 ENST00000405312.8 ENST00000451895.1 ENST00000433626.6 |
PSG4
|
pregnancy specific beta-1-glycoprotein 4 |
| chr10_-_30999469 | 0.34 |
ENST00000538351.6
|
ZNF438
|
zinc finger protein 438 |
| chr16_-_20900319 | 0.33 |
ENST00000564349.5
ENST00000324344.9 |
ERI2
DCUN1D3
|
ERI1 exoribonuclease family member 2 defective in cullin neddylation 1 domain containing 3 |
| chr5_-_151686908 | 0.33 |
ENST00000231061.9
|
SPARC
|
secreted protein acidic and cysteine rich |
| chr17_+_41226648 | 0.33 |
ENST00000377721.3
|
KRTAP9-2
|
keratin associated protein 9-2 |
| chr1_+_103571077 | 0.33 |
ENST00000610648.1
|
AMY2B
|
amylase alpha 2B |
| chr10_+_80132591 | 0.32 |
ENST00000372267.6
|
PLAC9
|
placenta associated 9 |
| chrX_+_9543103 | 0.31 |
ENST00000683056.1
|
TBL1X
|
transducin beta like 1 X-linked |
| chr1_+_103655760 | 0.30 |
ENST00000370083.9
|
AMY1A
|
amylase alpha 1A |
| chr1_+_103745323 | 0.29 |
ENST00000684141.1
|
AMY1C
|
amylase alpha 1C |
| chr12_+_26195543 | 0.29 |
ENST00000242729.7
|
SSPN
|
sarcospan |
| chr9_+_89605004 | 0.29 |
ENST00000252506.11
ENST00000375769.1 |
GADD45G
|
growth arrest and DNA damage inducible gamma |
| chr22_+_23856962 | 0.28 |
ENST00000611880.4
|
SLC2A11
|
solute carrier family 2 member 11 |
| chr1_-_48400826 | 0.28 |
ENST00000371841.1
|
SPATA6
|
spermatogenesis associated 6 |
| chr22_+_23856857 | 0.27 |
ENST00000403208.7
ENST00000398356.6 |
SLC2A11
|
solute carrier family 2 member 11 |
| chr7_+_74028066 | 0.27 |
ENST00000431562.5
ENST00000320492.11 ENST00000438906.5 |
ELN
|
elastin |
| chr11_+_20022550 | 0.27 |
ENST00000533917.5
|
NAV2
|
neuron navigator 2 |
| chr10_-_37858037 | 0.26 |
ENST00000395873.7
ENST00000357328.8 ENST00000395874.2 |
ZNF248
|
zinc finger protein 248 |
| chr2_-_85410336 | 0.26 |
ENST00000263867.9
ENST00000409921.5 |
CAPG
|
capping actin protein, gelsolin like |
| chr10_+_88880236 | 0.26 |
ENST00000371926.8
|
STAMBPL1
|
STAM binding protein like 1 |
| chr19_+_18386150 | 0.26 |
ENST00000252809.3
|
GDF15
|
growth differentiation factor 15 |
| chr11_-_96343170 | 0.26 |
ENST00000524717.6
|
MAML2
|
mastermind like transcriptional coactivator 2 |
| chr7_-_23470469 | 0.26 |
ENST00000258729.8
|
IGF2BP3
|
insulin like growth factor 2 mRNA binding protein 3 |
| chr12_+_26195313 | 0.26 |
ENST00000422622.3
|
SSPN
|
sarcospan |
| chr19_-_31349408 | 0.26 |
ENST00000240587.5
|
TSHZ3
|
teashirt zinc finger homeobox 3 |
| chr10_+_113709261 | 0.25 |
ENST00000672138.1
ENST00000452490.3 |
CASP7
|
caspase 7 |
| chr9_+_2159672 | 0.25 |
ENST00000634343.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr7_+_120988683 | 0.25 |
ENST00000340646.9
ENST00000310396.10 |
CPED1
|
cadherin like and PC-esterase domain containing 1 |
| chr21_-_26967057 | 0.25 |
ENST00000284987.6
|
ADAMTS5
|
ADAM metallopeptidase with thrombospondin type 1 motif 5 |
| chr7_+_74028127 | 0.25 |
ENST00000438880.5
ENST00000414324.5 ENST00000380562.8 ENST00000380575.8 ENST00000380584.8 ENST00000458204.5 ENST00000357036.9 ENST00000417091.5 ENST00000429192.5 ENST00000252034.12 ENST00000442310.5 ENST00000380553.8 ENST00000380576.9 ENST00000428787.5 ENST00000320399.10 |
ELN
|
elastin |
| chr12_-_71157992 | 0.25 |
ENST00000247829.8
|
TSPAN8
|
tetraspanin 8 |
| chr12_-_71157872 | 0.24 |
ENST00000546561.2
|
TSPAN8
|
tetraspanin 8 |
| chr10_+_103277129 | 0.24 |
ENST00000369849.9
|
INA
|
internexin neuronal intermediate filament protein alpha |
| chrX_+_136169891 | 0.24 |
ENST00000449474.5
|
FHL1
|
four and a half LIM domains 1 |
| chr22_+_23856703 | 0.24 |
ENST00000345044.10
|
SLC2A11
|
solute carrier family 2 member 11 |
| chr4_-_82844418 | 0.24 |
ENST00000503937.5
|
SEC31A
|
SEC31 homolog A, COPII coat complex component |
| chr14_-_106658251 | 0.24 |
ENST00000454421.2
|
IGHV3-64
|
immunoglobulin heavy variable 3-64 |
| chr10_+_89327989 | 0.23 |
ENST00000679923.1
ENST00000680085.1 ENST00000371818.9 ENST00000680779.1 |
IFIT3
|
interferon induced protein with tetratricopeptide repeats 3 |
| chr9_+_2159850 | 0.23 |
ENST00000416751.2
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr10_+_89327977 | 0.23 |
ENST00000681277.1
|
IFIT3
|
interferon induced protein with tetratricopeptide repeats 3 |
| chrX_-_63785510 | 0.23 |
ENST00000437457.6
ENST00000374878.5 ENST00000623517.3 |
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor 9 |
| chrX_+_56729231 | 0.23 |
ENST00000637096.1
ENST00000374922.9 ENST00000423617.2 |
NBDY
|
negative regulator of P-body association |
| chr3_-_71583683 | 0.23 |
ENST00000649631.1
ENST00000648718.1 |
FOXP1
|
forkhead box P1 |
| chr3_+_46354072 | 0.23 |
ENST00000445132.3
ENST00000421659.1 |
CCR2
|
C-C motif chemokine receptor 2 |
| chr9_+_121299793 | 0.22 |
ENST00000373818.8
|
GSN
|
gelsolin |
| chrX_+_136169833 | 0.22 |
ENST00000628032.2
|
FHL1
|
four and a half LIM domains 1 |
| chr15_-_89815332 | 0.21 |
ENST00000559874.2
|
ANPEP
|
alanyl aminopeptidase, membrane |
| chrX_+_136148440 | 0.21 |
ENST00000627383.2
ENST00000630084.2 |
FHL1
|
four and a half LIM domains 1 |
| chr10_-_125816596 | 0.20 |
ENST00000368786.5
|
UROS
|
uroporphyrinogen III synthase |
| chr14_+_23377136 | 0.20 |
ENST00000382809.2
|
CMTM5
|
CKLF like MARVEL transmembrane domain containing 5 |
| chr7_-_138798188 | 0.20 |
ENST00000310018.7
|
ATP6V0A4
|
ATPase H+ transporting V0 subunit a4 |
| chr7_-_138798104 | 0.20 |
ENST00000353492.4
|
ATP6V0A4
|
ATPase H+ transporting V0 subunit a4 |
| chr12_+_26195647 | 0.20 |
ENST00000535504.1
|
SSPN
|
sarcospan |
| chr20_+_15196834 | 0.20 |
ENST00000402914.5
|
MACROD2
|
mono-ADP ribosylhydrolase 2 |
| chr7_+_74027770 | 0.20 |
ENST00000445912.5
ENST00000621115.4 |
ELN
|
elastin |
| chr1_+_100719734 | 0.20 |
ENST00000370119.8
ENST00000294728.7 ENST00000347652.6 ENST00000370115.1 |
VCAM1
|
vascular cell adhesion molecule 1 |
| chr9_-_127905313 | 0.19 |
ENST00000622357.5
|
ST6GALNAC6
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chrX_+_136169664 | 0.19 |
ENST00000456445.5
|
FHL1
|
four and a half LIM domains 1 |
| chr3_-_122022122 | 0.19 |
ENST00000393631.5
ENST00000273691.7 ENST00000344209.10 |
ILDR1
|
immunoglobulin like domain containing receptor 1 |
| chr1_+_10430384 | 0.19 |
ENST00000470413.6
ENST00000602787.6 ENST00000309048.8 |
CENPS-CORT
CENPS
|
CENPS-CORT readthrough centromere protein S |
| chr6_+_36871841 | 0.19 |
ENST00000359359.6
|
C6orf89
|
chromosome 6 open reading frame 89 |
| chr8_-_132625378 | 0.19 |
ENST00000522789.5
|
LRRC6
|
leucine rich repeat containing 6 |
| chr2_-_127675459 | 0.19 |
ENST00000355119.9
|
LIMS2
|
LIM zinc finger domain containing 2 |
| chr3_-_71583592 | 0.19 |
ENST00000650156.1
ENST00000649596.1 |
FOXP1
|
forkhead box P1 |
| chr12_-_104050112 | 0.18 |
ENST00000547583.1
ENST00000546851.1 ENST00000360814.9 |
GLT8D2
|
glycosyltransferase 8 domain containing 2 |
| chr10_+_61662921 | 0.18 |
ENST00000648843.3
ENST00000330194.2 ENST00000389639.3 |
CABCOCO1
|
ciliary associated calcium binding coiled-coil 1 |
| chr12_-_91179517 | 0.18 |
ENST00000551354.1
|
DCN
|
decorin |
| chr5_-_134371004 | 0.17 |
ENST00000521755.1
ENST00000523054.5 ENST00000518409.1 |
CDKL3
ENSG00000273345.5
|
cyclin dependent kinase like 3 novel transcript |
| chr9_+_72628020 | 0.17 |
ENST00000646619.1
|
TMC1
|
transmembrane channel like 1 |
| chr10_-_7410973 | 0.17 |
ENST00000684547.1
ENST00000673876.1 ENST00000397167.6 |
SFMBT2
|
Scm like with four mbt domains 2 |
| chr21_-_10649835 | 0.17 |
ENST00000622028.1
|
IGHV1OR21-1
|
immunoglobulin heavy variable 1/OR21-1 (non-functional) |
| chr18_-_49813869 | 0.16 |
ENST00000586485.5
ENST00000587994.5 ENST00000586100.1 |
ACAA2
|
acetyl-CoA acyltransferase 2 |
| chr14_-_106235582 | 0.16 |
ENST00000390607.2
|
IGHV3-21
|
immunoglobulin heavy variable 3-21 |
| chr2_-_127675065 | 0.16 |
ENST00000545738.6
ENST00000409808.6 |
LIMS2
|
LIM zinc finger domain containing 2 |
| chr13_+_48976597 | 0.16 |
ENST00000541916.5
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr19_+_30372364 | 0.16 |
ENST00000355537.4
|
ZNF536
|
zinc finger protein 536 |
| chr12_+_103587266 | 0.16 |
ENST00000388887.7
|
STAB2
|
stabilin 2 |
| chr19_-_57559833 | 0.16 |
ENST00000457177.5
|
ZNF550
|
zinc finger protein 550 |
| chr1_+_171248471 | 0.16 |
ENST00000402921.6
ENST00000617670.6 ENST00000367750.7 |
FMO1
|
flavin containing dimethylaniline monoxygenase 1 |
| chr1_-_68232539 | 0.16 |
ENST00000370976.7
ENST00000354777.6 |
WLS
|
Wnt ligand secretion mediator |
| chr10_-_102432565 | 0.16 |
ENST00000369937.5
|
CUEDC2
|
CUE domain containing 2 |
| chr7_-_139716980 | 0.16 |
ENST00000342645.7
|
HIPK2
|
homeodomain interacting protein kinase 2 |
| chr1_-_15524183 | 0.16 |
ENST00000333868.10
ENST00000440484.1 |
CASP9
|
caspase 9 |
| chr5_+_141245384 | 0.16 |
ENST00000623671.1
ENST00000231173.6 |
PCDHB15
|
protocadherin beta 15 |
| chrX_+_136169624 | 0.15 |
ENST00000394153.6
|
FHL1
|
four and a half LIM domains 1 |
| chr20_-_58515382 | 0.15 |
ENST00000371149.8
|
APCDD1L
|
APC down-regulated 1 like |
| chr4_-_87391149 | 0.15 |
ENST00000507286.1
ENST00000358290.9 |
HSD17B11
|
hydroxysteroid 17-beta dehydrogenase 11 |
| chr3_-_71583713 | 0.15 |
ENST00000649528.3
ENST00000471386.3 ENST00000493089.7 |
FOXP1
|
forkhead box P1 |
| chr19_+_18340581 | 0.15 |
ENST00000604499.6
ENST00000269919.11 ENST00000595066.5 ENST00000252813.5 |
PGPEP1
|
pyroglutamyl-peptidase I |
| chr4_+_147732070 | 0.15 |
ENST00000336498.8
|
ARHGAP10
|
Rho GTPase activating protein 10 |
| chr15_+_84603896 | 0.15 |
ENST00000541040.5
ENST00000538076.5 ENST00000485222.2 |
ZSCAN2
|
zinc finger and SCAN domain containing 2 |
| chr4_+_26320975 | 0.15 |
ENST00000509158.6
ENST00000681856.1 ENST00000680140.1 ENST00000355476.8 ENST00000680511.1 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr16_-_21278282 | 0.15 |
ENST00000572914.2
|
CRYM
|
crystallin mu |
| chr9_-_90642855 | 0.15 |
ENST00000637905.1
|
DIRAS2
|
DIRAS family GTPase 2 |
| chr6_+_52423680 | 0.14 |
ENST00000538167.2
|
EFHC1
|
EF-hand domain containing 1 |
| chr15_-_19988117 | 0.14 |
ENST00000558565.2
|
IGHV3OR15-7
|
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr14_+_57268963 | 0.14 |
ENST00000261558.8
|
AP5M1
|
adaptor related protein complex 5 subunit mu 1 |
| chr1_+_152878312 | 0.14 |
ENST00000368765.4
|
SMCP
|
sperm mitochondria associated cysteine rich protein |
| chr12_+_59664677 | 0.14 |
ENST00000548610.5
|
SLC16A7
|
solute carrier family 16 member 7 |
| chr11_+_72192126 | 0.14 |
ENST00000393676.5
|
FOLR1
|
folate receptor alpha |
| chr1_+_53014926 | 0.14 |
ENST00000430330.6
ENST00000408941.7 ENST00000478274.6 ENST00000484100.5 ENST00000435345.6 ENST00000488965.1 |
SCP2
|
sterol carrier protein 2 |
| chr7_-_29195186 | 0.14 |
ENST00000449801.5
ENST00000409850.5 |
CPVL
|
carboxypeptidase vitellogenic like |
| chr22_-_35824373 | 0.14 |
ENST00000473487.6
|
RBFOX2
|
RNA binding fox-1 homolog 2 |
| chr20_+_58891302 | 0.14 |
ENST00000371095.7
ENST00000265620.11 ENST00000354359.12 ENST00000371085.8 |
GNAS
|
GNAS complex locus |
| chr5_+_176388731 | 0.14 |
ENST00000274787.3
|
HIGD2A
|
HIG1 hypoxia inducible domain family member 2A |
| chr3_+_160842143 | 0.14 |
ENST00000464260.5
ENST00000295839.9 |
PPM1L
|
protein phosphatase, Mg2+/Mn2+ dependent 1L |
| chr14_-_35121950 | 0.14 |
ENST00000554361.5
ENST00000261475.10 |
PPP2R3C
|
protein phosphatase 2 regulatory subunit B''gamma |
| chr14_+_44962177 | 0.13 |
ENST00000361462.7
ENST00000361577.7 |
TOGARAM1
|
TOG array regulator of axonemal microtubules 1 |
| chr10_-_110918934 | 0.13 |
ENST00000605742.5
|
BBIP1
|
BBSome interacting protein 1 |
| chr9_-_91423819 | 0.13 |
ENST00000297689.4
|
NFIL3
|
nuclear factor, interleukin 3 regulated |
| chr10_-_110919146 | 0.13 |
ENST00000652396.1
ENST00000423273.5 ENST00000436562.1 ENST00000651952.1 ENST00000448814.7 ENST00000652400.1 ENST00000447005.5 ENST00000454061.5 |
BBIP1
|
BBSome interacting protein 1 |
| chr10_+_110919595 | 0.13 |
ENST00000369452.9
|
SHOC2
|
SHOC2 leucine rich repeat scaffold protein |
| chr12_-_49187369 | 0.13 |
ENST00000547939.6
|
TUBA1A
|
tubulin alpha 1a |
| chr17_-_38748184 | 0.13 |
ENST00000618941.4
ENST00000620225.5 ENST00000618506.1 ENST00000616129.4 |
PCGF2
|
polycomb group ring finger 2 |
| chr4_+_26320782 | 0.12 |
ENST00000514807.5
ENST00000348160.9 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr1_+_61081728 | 0.12 |
ENST00000371189.8
|
NFIA
|
nuclear factor I A |
| chr16_-_29899245 | 0.12 |
ENST00000537485.5
|
SEZ6L2
|
seizure related 6 homolog like 2 |
| chr1_-_15524344 | 0.12 |
ENST00000348549.9
ENST00000546424.5 |
CASP9
|
caspase 9 |
| chr10_+_80132722 | 0.12 |
ENST00000372263.4
|
PLAC9
|
placenta associated 9 |
| chr14_-_106117159 | 0.12 |
ENST00000390601.3
|
IGHV3-11
|
immunoglobulin heavy variable 3-11 |
| chr12_-_123436664 | 0.12 |
ENST00000280571.10
|
RILPL2
|
Rab interacting lysosomal protein like 2 |
| chr10_+_117543567 | 0.12 |
ENST00000616794.1
|
EMX2
|
empty spiracles homeobox 2 |
| chr4_+_133149307 | 0.12 |
ENST00000618019.1
|
PCDH10
|
protocadherin 10 |
| chr3_-_183099508 | 0.12 |
ENST00000476176.5
|
MCCC1
|
methylcrotonoyl-CoA carboxylase 1 |
| chr11_+_61755372 | 0.12 |
ENST00000265460.9
|
MYRF
|
myelin regulatory factor |
| chr16_-_29899043 | 0.12 |
ENST00000346932.9
ENST00000350527.7 ENST00000568380.1 |
SEZ6L2
|
seizure related 6 homolog like 2 |
| chr3_-_158105718 | 0.12 |
ENST00000554685.2
|
SHOX2
|
short stature homeobox 2 |
| chrX_-_15601077 | 0.12 |
ENST00000680121.1
|
ACE2
|
angiotensin I converting enzyme 2 |
| chr6_+_68635273 | 0.12 |
ENST00000370598.6
|
ADGRB3
|
adhesion G protein-coupled receptor B3 |
| chr11_-_102724945 | 0.12 |
ENST00000236826.8
|
MMP8
|
matrix metallopeptidase 8 |
| chr1_-_47190013 | 0.11 |
ENST00000294338.7
|
PDZK1IP1
|
PDZK1 interacting protein 1 |
| chr2_+_63842325 | 0.11 |
ENST00000445915.6
ENST00000475462.5 |
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr4_+_26320563 | 0.11 |
ENST00000361572.10
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr14_+_64503943 | 0.11 |
ENST00000556965.1
ENST00000554015.5 |
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr1_-_159714581 | 0.11 |
ENST00000255030.9
ENST00000437342.1 ENST00000368112.5 ENST00000368111.5 ENST00000368110.1 |
CRP
|
C-reactive protein |
| chr5_+_43602648 | 0.11 |
ENST00000505678.6
ENST00000512422.5 ENST00000264663.9 ENST00000670904.1 ENST00000653251.1 |
NNT
|
nicotinamide nucleotide transhydrogenase |
Network of associatons between targets according to the STRING database.
First level regulatory network of IKZF2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.7 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.4 | 2.5 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.3 | 1.3 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.3 | 1.2 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.3 | 1.7 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.3 | 0.8 | GO:0035691 | macrophage migration inhibitory factor signaling pathway(GO:0035691) |
| 0.2 | 4.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.2 | 0.7 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 2.3 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.1 | 0.6 | GO:0034444 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.1 | 0.4 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.1 | 0.3 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.1 | 2.0 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.1 | 0.4 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.1 | 1.6 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.1 | 0.6 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 0.4 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.1 | 0.8 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 0.2 | GO:0010607 | negative regulation of cytoplasmic mRNA processing body assembly(GO:0010607) |
| 0.1 | 0.2 | GO:1903977 | immune complex clearance(GO:0002434) immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) negative regulation of eosinophil activation(GO:1902567) positive regulation of glial cell migration(GO:1903977) positive regulation of monocyte extravasation(GO:2000439) |
| 0.1 | 0.2 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 0.5 | GO:2000343 | positive regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000343) |
| 0.1 | 0.3 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.1 | 0.4 | GO:1905068 | positive regulation of ephrin receptor signaling pathway(GO:1901189) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.1 | 0.2 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.1 | 0.2 | GO:1903259 | exon-exon junction complex disassembly(GO:1903259) |
| 0.0 | 0.1 | GO:0003147 | neural crest cell migration involved in heart formation(GO:0003147) cell migration involved in heart formation(GO:0060974) anterior neural tube closure(GO:0061713) |
| 0.0 | 0.3 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) vagus nerve development(GO:0021564) |
| 0.0 | 1.0 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.2 | GO:0044858 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.7 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.5 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.1 | GO:2000174 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.0 | 0.3 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.4 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.1 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.0 | 0.1 | GO:0070846 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 | 0.6 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.3 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.5 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 1.1 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.8 | GO:0071380 | cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.0 | 0.1 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.3 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.4 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) regulation of bleb assembly(GO:1904170) positive regulation of bleb assembly(GO:1904172) |
| 0.0 | 0.2 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.2 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.4 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 3.1 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.2 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.1 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.5 | GO:0007351 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 | 0.4 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.9 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.2 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.0 | 0.1 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.3 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.2 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.2 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.1 | GO:0019255 | UDP-glucuronate biosynthetic process(GO:0006065) glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.0 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.3 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.5 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.5 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.1 | GO:0060849 | radial pattern formation(GO:0009956) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.0 | 0.2 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.2 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.8 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.9 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.0 | GO:2000724 | mineralocorticoid receptor signaling pathway(GO:0031959) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.0 | 0.1 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.1 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.0 | 0.3 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.2 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.2 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.3 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.1 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.0 | 0.3 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.1 | GO:0043366 | beta selection(GO:0043366) |
| 0.0 | 0.0 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.3 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 1.0 | GO:0034113 | heterotypic cell-cell adhesion(GO:0034113) |
| 0.0 | 0.1 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.1 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.0 | GO:0048880 | thiamine pyrophosphate transport(GO:0030974) sensory system development(GO:0048880) |
| 0.0 | 0.0 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.3 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.4 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.9 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 0.2 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) kinetochore assembly(GO:0051382) |
| 0.0 | 0.1 | GO:0042262 | DNA protection(GO:0042262) |
| 0.0 | 0.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.8 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.4 | 4.9 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.3 | 4.2 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.6 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.1 | 0.3 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 0.3 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.1 | 0.7 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 0.2 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.0 | 0.2 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.4 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.1 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 0.0 | 0.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 1.4 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 1.1 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.2 | GO:1905202 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.0 | 0.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 1.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.3 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.3 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.3 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 1.5 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 2.3 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.0 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.4 | 2.3 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.3 | 4.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 2.5 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.2 | 0.6 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.2 | 0.5 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.2 | 0.5 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.1 | 0.4 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.1 | 0.4 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.1 | 0.4 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 0.2 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 1.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.2 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 1.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.2 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.1 | 4.7 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.4 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.8 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.5 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.1 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.1 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.0 | 0.9 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.6 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.1 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.3 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.4 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.2 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.0 | 0.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 1.0 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.2 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.0 | GO:0030305 | heparanase activity(GO:0030305) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.4 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.4 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.1 | GO:0030614 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) |
| 0.0 | 0.4 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.4 | GO:0044769 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.8 | GO:0051119 | sugar transmembrane transporter activity(GO:0051119) |
| 0.0 | 0.0 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.4 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.0 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.2 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 1.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 0.0 | GO:0090422 | thiamine pyrophosphate transporter activity(GO:0090422) |
| 0.0 | 0.2 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 1.2 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 1.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 3.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.8 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 2.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.8 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 2.5 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.0 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.2 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 0.9 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 1.8 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 1.7 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.1 | 1.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 3.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.8 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.7 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 2.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.6 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.8 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |