|
chr6_+_72212887
Show fit
|
0.91 |
ENST00000523963.5
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr6_+_72212802
Show fit
|
0.90 |
ENST00000401910.7
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr11_+_114060204
Show fit
|
0.88 |
ENST00000683318.1
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr17_+_6756035
Show fit
|
0.72 |
ENST00000361842.8
ENST00000574907.5
|
XAF1
|
XIAP associated factor 1
|
|
chr17_+_6755834
Show fit
|
0.67 |
ENST00000346752.8
|
XAF1
|
XIAP associated factor 1
|
|
chr13_-_42992165
Show fit
|
0.64 |
ENST00000398762.7
ENST00000313640.11
ENST00000313624.12
|
EPSTI1
|
epithelial stromal interaction 1
|
|
chr5_+_32710630
Show fit
|
0.64 |
ENST00000326958.5
|
NPR3
|
natriuretic peptide receptor 3
|
|
chr1_+_78649818
Show fit
|
0.57 |
ENST00000370747.9
ENST00000438486.1
|
IFI44
|
interferon induced protein 44
|
|
chr18_-_28036585
Show fit
|
0.55 |
ENST00000399380.7
|
CDH2
|
cadherin 2
|
|
chr1_+_162632454
Show fit
|
0.54 |
ENST00000367921.8
ENST00000367922.7
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr6_-_81752671
Show fit
|
0.45 |
ENST00000320172.11
ENST00000369754.7
ENST00000369756.3
|
TENT5A
|
terminal nucleotidyltransferase 5A
|
|
chr1_+_220879434
Show fit
|
0.44 |
ENST00000366903.8
|
HLX
|
H2.0 like homeobox
|
|
chr3_-_122564232
Show fit
|
0.41 |
ENST00000471785.5
ENST00000466126.1
|
PARP9
|
poly(ADP-ribose) polymerase family member 9
|
|
chr14_+_24161257
Show fit
|
0.41 |
ENST00000396864.8
ENST00000557894.5
ENST00000559284.5
ENST00000560275.5
|
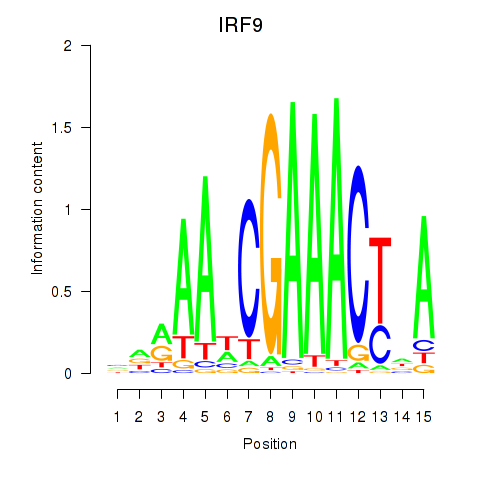
IRF9
|
interferon regulatory factor 9
|
|
chr3_-_122564577
Show fit
|
0.39 |
ENST00000477522.6
ENST00000360356.6
|
PARP9
|
poly(ADP-ribose) polymerase family member 9
|
|
chr11_-_86672114
Show fit
|
0.34 |
ENST00000393324.7
|
ME3
|
malic enzyme 3
|
|
chr11_-_86672630
Show fit
|
0.33 |
ENST00000543262.5
|
ME3
|
malic enzyme 3
|
|
chr11_-_86672419
Show fit
|
0.32 |
ENST00000524826.7
ENST00000532471.1
|
ME3
|
malic enzyme 3
|
|
chr11_-_57567617
Show fit
|
0.31 |
ENST00000287156.9
|
UBE2L6
|
ubiquitin conjugating enzyme E2 L6
|
|
chr7_+_134866831
Show fit
|
0.30 |
ENST00000435928.1
|
CALD1
|
caldesmon 1
|
|
chr2_-_151289613
Show fit
|
0.28 |
ENST00000243346.10
|
NMI
|
N-myc and STAT interactor
|
|
chr15_+_91100194
Show fit
|
0.28 |
ENST00000394232.6
|
SV2B
|
synaptic vesicle glycoprotein 2B
|
|
chr1_+_1013485
Show fit
|
0.27 |
ENST00000649529.1
|
ISG15
|
ISG15 ubiquitin like modifier
|
|
chr9_-_92536031
Show fit
|
0.27 |
ENST00000344604.9
ENST00000375540.5
|
ECM2
|
extracellular matrix protein 2
|
|
chr4_+_186069144
Show fit
|
0.27 |
ENST00000513189.1
ENST00000296795.8
|
TLR3
|
toll like receptor 3
|
|
chr10_+_89332484
Show fit
|
0.26 |
ENST00000371811.4
ENST00000680037.1
ENST00000679583.1
ENST00000679897.1
|
IFIT3
|
interferon induced protein with tetratricopeptide repeats 3
|
|
chr3_-_121660892
Show fit
|
0.26 |
ENST00000428394.6
ENST00000314583.8
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr19_-_17405554
Show fit
|
0.23 |
ENST00000252593.7
|
BST2
|
bone marrow stromal cell antigen 2
|
|
chr3_+_187368367
Show fit
|
0.22 |
ENST00000259030.3
|
RTP4
|
receptor transporter protein 4
|
|
chrX_-_63785510
Show fit
|
0.22 |
ENST00000437457.6
ENST00000374878.5
ENST00000623517.3
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor 9
|
|
chr4_+_159241016
Show fit
|
0.21 |
ENST00000644902.1
|
RAPGEF2
|
Rap guanine nucleotide exchange factor 2
|
|
chr6_-_24935942
Show fit
|
0.20 |
ENST00000645100.1
ENST00000643898.2
ENST00000613507.4
|
RIPOR2
|
RHO family interacting cell polarization regulator 2
|
|
chr17_+_48831021
Show fit
|
0.20 |
ENST00000509507.5
ENST00000448105.6
ENST00000570513.5
ENST00000509415.5
ENST00000513119.5
ENST00000416445.6
ENST00000508679.5
ENST00000258947.8
ENST00000505071.5
|
CALCOCO2
|
calcium binding and coiled-coil domain 2
|
|
chr10_+_89414555
Show fit
|
0.20 |
ENST00000371795.5
ENST00000681422.1
|
IFIT5
|
interferon induced protein with tetratricopeptide repeats 5
|
|
chr7_+_101085464
Show fit
|
0.19 |
ENST00000306085.11
ENST00000412507.1
|
TRIM56
|
tripartite motif containing 56
|
|
chr17_+_80260826
Show fit
|
0.19 |
ENST00000508628.6
ENST00000582970.5
ENST00000319921.4
|
RNF213
|
ring finger protein 213
|
|
chr1_+_241532121
Show fit
|
0.18 |
ENST00000366558.7
|
KMO
|
kynurenine 3-monooxygenase
|
|
chr10_+_89301932
Show fit
|
0.18 |
ENST00000371826.4
ENST00000679755.1
|
IFIT2
|
interferon induced protein with tetratricopeptide repeats 2
|
|
chr14_+_100065400
Show fit
|
0.17 |
ENST00000555706.5
ENST00000392920.8
ENST00000555048.5
|
EVL
|
Enah/Vasp-like
|
|
chr9_-_21995301
Show fit
|
0.16 |
ENST00000498628.6
|
CDKN2A
|
cyclin dependent kinase inhibitor 2A
|
|
chr16_-_15643024
Show fit
|
0.16 |
ENST00000540441.6
|
MARF1
|
meiosis regulator and mRNA stability factor 1
|
|
chr3_-_179451387
Show fit
|
0.15 |
ENST00000675901.1
ENST00000232564.8
ENST00000674862.1
ENST00000497513.1
|
GNB4
|
G protein subunit beta 4
|
|
chr11_-_615570
Show fit
|
0.15 |
ENST00000649187.1
ENST00000647801.1
ENST00000397566.5
ENST00000397570.5
|
IRF7
|
interferon regulatory factor 7
|
|
chr4_-_76213724
Show fit
|
0.15 |
ENST00000639738.1
|
SCARB2
|
scavenger receptor class B member 2
|
|
chr4_-_76213846
Show fit
|
0.15 |
ENST00000639145.1
|
SCARB2
|
scavenger receptor class B member 2
|
|
chr3_-_146544538
Show fit
|
0.13 |
ENST00000462666.5
|
PLSCR1
|
phospholipid scramblase 1
|
|
chr15_+_73994694
Show fit
|
0.13 |
ENST00000268058.8
ENST00000395132.6
ENST00000268059.10
ENST00000354026.10
ENST00000565898.5
ENST00000569477.5
ENST00000569965.5
ENST00000567543.5
ENST00000436891.7
ENST00000435786.6
ENST00000564428.5
ENST00000359928.8
|
PML
|
PML nuclear body scaffold
|
|
chr3_-_146544850
Show fit
|
0.13 |
ENST00000472349.1
|
PLSCR1
|
phospholipid scramblase 1
|
|
chr2_-_151261839
Show fit
|
0.13 |
ENST00000331426.6
|
RBM43
|
RNA binding motif protein 43
|
|
chr9_-_21995262
Show fit
|
0.13 |
ENST00000494262.5
|
CDKN2A
|
cyclin dependent kinase inhibitor 2A
|
|
chr17_-_4263847
Show fit
|
0.13 |
ENST00000570535.5
ENST00000574367.5
ENST00000341657.9
|
ANKFY1
|
ankyrin repeat and FYVE domain containing 1
|
|
chr4_-_76213817
Show fit
|
0.12 |
ENST00000264896.8
ENST00000640640.1
ENST00000640957.1
|
SCARB2
|
scavenger receptor class B member 2
|
|
chr6_+_125919296
Show fit
|
0.12 |
ENST00000444128.2
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr3_-_146544636
Show fit
|
0.12 |
ENST00000486631.5
|
PLSCR1
|
phospholipid scramblase 1
|
|
chr6_+_3068487
Show fit
|
0.12 |
ENST00000259808.9
|
RIPK1
|
receptor interacting serine/threonine kinase 1
|
|
chr4_-_76213589
Show fit
|
0.12 |
ENST00000638603.1
ENST00000452464.6
|
SCARB2
|
scavenger receptor class B member 2
|
|
chr12_-_121039204
Show fit
|
0.12 |
ENST00000620239.5
|
OASL
|
2'-5'-oligoadenylate synthetase like
|
|
chr14_+_22462932
Show fit
|
0.12 |
ENST00000390477.2
|
TRDC
|
T cell receptor delta constant
|
|
chr12_-_56360084
Show fit
|
0.12 |
ENST00000314128.9
ENST00000557235.5
ENST00000651915.1
|
STAT2
|
signal transducer and activator of transcription 2
|
|
chr3_-_146544701
Show fit
|
0.12 |
ENST00000487389.5
|
PLSCR1
|
phospholipid scramblase 1
|
|
chr1_+_154405193
Show fit
|
0.12 |
ENST00000622330.4
ENST00000344086.8
|
IL6R
|
interleukin 6 receptor
|
|
chr16_+_28863812
Show fit
|
0.11 |
ENST00000684370.1
|
SH2B1
|
SH2B adaptor protein 1
|
|
chr6_+_125919210
Show fit
|
0.11 |
ENST00000438495.6
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr12_-_121039236
Show fit
|
0.11 |
ENST00000257570.9
|
OASL
|
2'-5'-oligoadenylate synthetase like
|
|
chr10_+_113679523
Show fit
|
0.11 |
ENST00000345633.8
ENST00000614447.4
ENST00000369321.6
|
CASP7
|
caspase 7
|
|
chr20_-_49278034
Show fit
|
0.11 |
ENST00000371744.5
ENST00000396105.6
ENST00000371752.5
|
ZNFX1
|
zinc finger NFX1-type containing 1
|
|
chr12_-_121039156
Show fit
|
0.11 |
ENST00000339275.10
|
OASL
|
2'-5'-oligoadenylate synthetase like
|
|
chr10_+_113679839
Show fit
|
0.11 |
ENST00000369318.8
ENST00000369315.5
|
CASP7
|
caspase 7
|
|
chr5_+_134526100
Show fit
|
0.11 |
ENST00000395003.5
|
JADE2
|
jade family PHD finger 2
|
|
chr11_+_67023085
Show fit
|
0.10 |
ENST00000527043.6
|
SYT12
|
synaptotagmin 12
|
|
chrX_-_49002197
Show fit
|
0.10 |
ENST00000622231.1
ENST00000617369.4
ENST00000593475.5
ENST00000622599.4
|
GRIPAP1
|
GRIP1 associated protein 1
|
|
chr3_+_122680802
Show fit
|
0.10 |
ENST00000474629.7
|
PARP14
|
poly(ADP-ribose) polymerase family member 14
|
|
chr19_+_17405685
Show fit
|
0.10 |
ENST00000646744.1
ENST00000635536.2
ENST00000635060.2
ENST00000634291.2
ENST00000645298.1
ENST00000528911.5
|
BISPR
MVB12A
|
BST2 interferon stimulated positive regulator
multivesicular body subunit 12A
|
|
chrX_-_49002256
Show fit
|
0.10 |
ENST00000611705.4
ENST00000376423.8
|
GRIPAP1
|
GRIP1 associated protein 1
|
|
chr3_-_146544578
Show fit
|
0.10 |
ENST00000342435.9
ENST00000448787.6
|
PLSCR1
|
phospholipid scramblase 1
|
|
chr3_-_122564253
Show fit
|
0.10 |
ENST00000492382.5
ENST00000682323.1
ENST00000462315.5
|
PARP9
|
poly(ADP-ribose) polymerase family member 9
|
|
chr11_-_615921
Show fit
|
0.09 |
ENST00000348655.11
ENST00000525445.6
ENST00000330243.9
|
IRF7
|
interferon regulatory factor 7
|
|
chr2_+_33476640
Show fit
|
0.09 |
ENST00000425210.5
ENST00000444784.5
ENST00000423159.5
ENST00000403687.8
|
RASGRP3
|
RAS guanyl releasing protein 3
|
|
chr3_-_28348805
Show fit
|
0.09 |
ENST00000457172.5
ENST00000479665.6
|
AZI2
|
5-azacytidine induced 2
|
|
chr1_-_182589239
Show fit
|
0.08 |
ENST00000367559.7
ENST00000539397.1
|
RNASEL
|
ribonuclease L
|
|
chr1_+_236524581
Show fit
|
0.08 |
ENST00000526634.5
|
LGALS8
|
galectin 8
|
|
chr6_-_117573571
Show fit
|
0.08 |
ENST00000467125.1
|
ENSG00000282218.1
|
novel protein, GOPC-ROS1 readthrough
|
|
chr6_-_110415539
Show fit
|
0.08 |
ENST00000368923.8
ENST00000368924.9
|
DDO
|
D-aspartate oxidase
|
|
chrX_-_40097403
Show fit
|
0.08 |
ENST00000397354.7
|
BCOR
|
BCL6 corepressor
|
|
chr13_+_49495941
Show fit
|
0.08 |
ENST00000378319.8
ENST00000496623.5
ENST00000426879.5
|
PHF11
|
PHD finger protein 11
|
|
chr3_-_71360753
Show fit
|
0.07 |
ENST00000648783.1
|
FOXP1
|
forkhead box P1
|
|
chr13_+_49496355
Show fit
|
0.07 |
ENST00000496612.5
ENST00000357596.7
ENST00000485919.5
ENST00000442195.5
|
PHF11
|
PHD finger protein 11
|
|
chr3_-_71581540
Show fit
|
0.07 |
ENST00000650068.1
|
FOXP1
|
forkhead box P1
|
|
chr1_-_206946448
Show fit
|
0.06 |
ENST00000356495.5
|
PIGR
|
polymeric immunoglobulin receptor
|
|
chr1_+_241532370
Show fit
|
0.06 |
ENST00000366559.9
ENST00000366557.8
|
KMO
|
kynurenine 3-monooxygenase
|
|
chr12_+_6772512
Show fit
|
0.06 |
ENST00000441671.6
ENST00000203629.3
|
LAG3
|
lymphocyte activating 3
|
|
chr12_-_121038967
Show fit
|
0.06 |
ENST00000680620.1
ENST00000679655.1
ENST00000543677.2
|
OASL
|
2'-5'-oligoadenylate synthetase like
|
|
chr17_+_27631148
Show fit
|
0.06 |
ENST00000313648.10
ENST00000395473.7
ENST00000577392.5
ENST00000584661.5
|
LGALS9
|
galectin 9
|
|
chr12_-_27014300
Show fit
|
0.06 |
ENST00000535819.1
ENST00000543803.5
ENST00000535423.5
ENST00000539741.5
ENST00000343028.9
ENST00000545600.1
ENST00000543088.5
|
TM7SF3
|
transmembrane 7 superfamily member 3
|
|
chr1_-_161631032
Show fit
|
0.06 |
ENST00000534776.1
ENST00000613418.4
ENST00000614870.4
|
FCGR3B
|
Fc fragment of IgG receptor IIIb
|
|
chr3_+_63911929
Show fit
|
0.06 |
ENST00000487717.5
|
ATXN7
|
ataxin 7
|
|
chr19_+_17406099
Show fit
|
0.05 |
ENST00000634942.2
ENST00000634568.1
ENST00000600514.5
|
BISPR
MVB12A
|
BST2 interferon stimulated positive regulator
multivesicular body subunit 12A
|
|
chr19_+_17405734
Show fit
|
0.05 |
ENST00000635572.1
ENST00000634675.1
ENST00000634813.1
ENST00000647283.1
ENST00000635435.2
ENST00000634731.2
ENST00000635339.1
ENST00000528604.5
|
BISPR
MVB12A
|
BST2 interferon stimulated positive regulator
multivesicular body subunit 12A
|
|
chr3_+_63912588
Show fit
|
0.05 |
ENST00000522345.2
|
ATXN7
|
ataxin 7
|
|
chr8_-_143986425
Show fit
|
0.05 |
ENST00000313059.9
ENST00000524918.5
ENST00000313028.12
ENST00000525773.5
|
PARP10
|
poly(ADP-ribose) polymerase family member 10
|
|
chr9_+_99906646
Show fit
|
0.05 |
ENST00000259400.11
ENST00000531035.5
ENST00000525640.5
ENST00000534052.1
ENST00000526607.1
|
STX17
|
syntaxin 17
|
|
chr20_+_62804794
Show fit
|
0.05 |
ENST00000290291.10
|
OGFR
|
opioid growth factor receptor
|
|
chr9_-_33264559
Show fit
|
0.04 |
ENST00000473781.1
ENST00000379704.7
ENST00000488499.1
|
BAG1
|
BAG cochaperone 1
|
|
chr1_-_161631152
Show fit
|
0.04 |
ENST00000421702.3
ENST00000650385.1
|
FCGR3B
|
Fc fragment of IgG receptor IIIb
|
|
chr3_-_28348924
Show fit
|
0.04 |
ENST00000414162.5
ENST00000420543.6
|
AZI2
|
5-azacytidine induced 2
|
|
chr4_+_37891060
Show fit
|
0.04 |
ENST00000261439.9
ENST00000508802.5
ENST00000402522.1
|
TBC1D1
|
TBC1 domain family member 1
|
|
chr22_+_18150162
Show fit
|
0.04 |
ENST00000215794.8
|
USP18
|
ubiquitin specific peptidase 18
|
|
chr6_-_105137147
Show fit
|
0.04 |
ENST00000314641.10
|
BVES
|
blood vessel epicardial substance
|
|
chr17_+_40140500
Show fit
|
0.03 |
ENST00000264645.12
|
CASC3
|
CASC3 exon junction complex subunit
|
|
chr18_-_69956924
Show fit
|
0.03 |
ENST00000581982.5
ENST00000280200.8
|
CD226
|
CD226 molecule
|
|
chr19_+_16143678
Show fit
|
0.03 |
ENST00000613986.4
ENST00000593031.1
|
HSH2D
|
hematopoietic SH2 domain containing
|
|
chr1_+_156082563
Show fit
|
0.03 |
ENST00000368301.6
|
LMNA
|
lamin A/C
|
|
chr13_-_40982880
Show fit
|
0.03 |
ENST00000635415.1
|
ELF1
|
E74 like ETS transcription factor 1
|
|
chr21_-_33931607
Show fit
|
0.03 |
ENST00000429238.2
|
ENSG00000249209.2
|
novel protein similar to ATP synthase delta (OSCP) subunit domain
|
|
chr3_-_28348629
Show fit
|
0.02 |
ENST00000334100.10
|
AZI2
|
5-azacytidine induced 2
|
|
chr20_+_11917859
Show fit
|
0.02 |
ENST00000618296.4
ENST00000378226.7
|
BTBD3
|
BTB domain containing 3
|
|
chr3_-_142028617
Show fit
|
0.02 |
ENST00000477292.5
ENST00000478006.5
ENST00000495310.5
ENST00000486111.5
|
TFDP2
|
transcription factor Dp-2
|
|
chr14_-_67515153
Show fit
|
0.02 |
ENST00000555994.6
|
TMEM229B
|
transmembrane protein 229B
|
|
chr3_-_123980727
Show fit
|
0.02 |
ENST00000620893.4
|
ROPN1
|
rhophilin associated tail protein 1
|
|
chr15_+_73994777
Show fit
|
0.02 |
ENST00000563500.5
|
PML
|
PML nuclear body scaffold
|
|
chr4_+_87006988
Show fit
|
0.01 |
ENST00000307808.10
|
AFF1
|
AF4/FMR2 family member 1
|
|
chr4_-_74794514
Show fit
|
0.01 |
ENST00000395743.8
|
BTC
|
betacellulin
|
|
chr1_+_159009886
Show fit
|
0.01 |
ENST00000340979.10
ENST00000368131.8
ENST00000295809.12
ENST00000368132.7
|
IFI16
|
interferon gamma inducible protein 16
|
|
chr6_-_33314247
Show fit
|
0.01 |
ENST00000475304.5
ENST00000489157.5
|
TAPBP
|
TAP binding protein
|
|
chr6_-_33314202
Show fit
|
0.01 |
ENST00000426633.6
ENST00000467025.1
|
TAPBP
|
TAP binding protein
|
|
chr17_-_42112674
Show fit
|
0.01 |
ENST00000251642.8
ENST00000591220.5
|
DHX58
|
DExH-box helicase 58
|
|
chr4_-_76213520
Show fit
|
0.00 |
ENST00000640634.1
|
SCARB2
|
scavenger receptor class B member 2
|
|
chr5_+_90640718
Show fit
|
0.00 |
ENST00000640403.1
|
ADGRV1
|
adhesion G protein-coupled receptor V1
|
|
chr4_-_168318743
Show fit
|
0.00 |
ENST00000393743.8
|
DDX60
|
DExD/H-box helicase 60
|
|
chr3_+_122564327
Show fit
|
0.00 |
ENST00000296161.9
ENST00000383661.3
|
DTX3L
|
deltex E3 ubiquitin ligase 3L
|
|
chr18_+_3262956
Show fit
|
0.00 |
ENST00000584539.1
|
MYL12B
|
myosin light chain 12B
|
|
chr9_+_68356603
Show fit
|
0.00 |
ENST00000396396.6
|
PGM5
|
phosphoglucomutase 5
|
|
chr11_+_5624987
Show fit
|
0.00 |
ENST00000429814.3
|
TRIM34
|
tripartite motif containing 34
|