Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
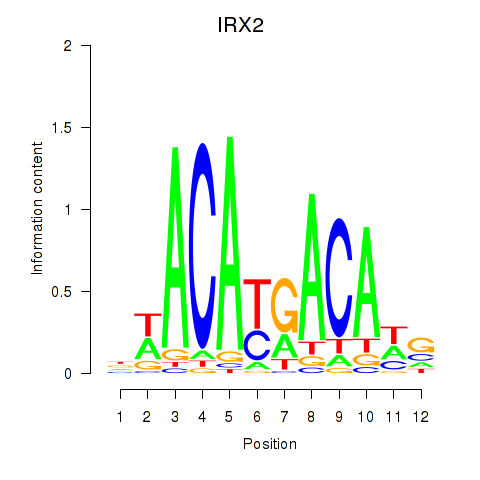
Results for IRX2
Z-value: 1.06
Transcription factors associated with IRX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRX2
|
ENSG00000170561.13 | iroquois homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRX2 | hg38_v1_chr5_-_2751648_2751668 | 0.62 | 1.0e-01 | Click! |
Activity profile of IRX2 motif
Sorted Z-values of IRX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_98268186 | 1.41 |
ENST00000260702.4
|
LOXL4
|
lysyl oxidase like 4 |
| chr16_+_68644988 | 1.08 |
ENST00000429102.6
|
CDH3
|
cadherin 3 |
| chr17_-_3691887 | 0.89 |
ENST00000552050.5
|
P2RX5
|
purinergic receptor P2X 5 |
| chr5_+_36608146 | 0.77 |
ENST00000381918.4
ENST00000513646.1 |
SLC1A3
|
solute carrier family 1 member 3 |
| chr11_-_7830840 | 0.75 |
ENST00000641167.1
|
OR5P3
|
olfactory receptor family 5 subfamily P member 3 |
| chr3_+_122325237 | 0.75 |
ENST00000264474.4
ENST00000479204.1 |
CSTA
|
cystatin A |
| chr3_-_112846856 | 0.66 |
ENST00000488794.5
|
CD200R1L
|
CD200 receptor 1 like |
| chr6_-_136525961 | 0.64 |
ENST00000438100.6
|
MAP7
|
microtubule associated protein 7 |
| chr6_-_136526177 | 0.63 |
ENST00000617204.4
|
MAP7
|
microtubule associated protein 7 |
| chr6_-_136526472 | 0.58 |
ENST00000454590.5
ENST00000432797.6 |
MAP7
|
microtubule associated protein 7 |
| chr6_-_136526654 | 0.54 |
ENST00000611373.1
|
MAP7
|
microtubule associated protein 7 |
| chr4_-_22443110 | 0.53 |
ENST00000508133.5
|
ADGRA3
|
adhesion G protein-coupled receptor A3 |
| chr9_-_137302264 | 0.50 |
ENST00000356628.4
|
NRARP
|
NOTCH regulated ankyrin repeat protein |
| chr8_-_85341705 | 0.48 |
ENST00000517618.5
|
CA1
|
carbonic anhydrase 1 |
| chr16_+_82035245 | 0.45 |
ENST00000199936.9
|
HSD17B2
|
hydroxysteroid 17-beta dehydrogenase 2 |
| chr11_+_62880863 | 0.44 |
ENST00000680297.1
|
SLC3A2
|
solute carrier family 3 member 2 |
| chr11_+_62880885 | 0.44 |
ENST00000541372.1
ENST00000539458.1 ENST00000338663.12 ENST00000681232.1 ENST00000681657.1 |
SLC3A2
|
solute carrier family 3 member 2 |
| chrX_+_136536099 | 0.42 |
ENST00000440515.5
ENST00000456412.1 |
VGLL1
|
vestigial like family member 1 |
| chr4_+_87975667 | 0.42 |
ENST00000237623.11
ENST00000682655.1 ENST00000508233.6 ENST00000360804.4 ENST00000395080.8 |
SPP1
|
secreted phosphoprotein 1 |
| chr1_+_31410164 | 0.40 |
ENST00000536859.5
|
SERINC2
|
serine incorporator 2 |
| chr17_-_27800524 | 0.39 |
ENST00000313735.11
|
NOS2
|
nitric oxide synthase 2 |
| chr18_-_55403682 | 0.38 |
ENST00000564228.5
ENST00000630828.2 |
TCF4
|
transcription factor 4 |
| chr5_+_147878703 | 0.36 |
ENST00000296694.5
|
SCGB3A2
|
secretoglobin family 3A member 2 |
| chr10_+_125973373 | 0.30 |
ENST00000417114.5
ENST00000445510.5 ENST00000368691.5 |
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr15_-_44194407 | 0.29 |
ENST00000484674.5
|
FRMD5
|
FERM domain containing 5 |
| chr8_-_85341659 | 0.29 |
ENST00000522389.5
|
CA1
|
carbonic anhydrase 1 |
| chr10_-_59362584 | 0.28 |
ENST00000618427.4
ENST00000611933.4 |
FAM13C
|
family with sequence similarity 13 member C |
| chr12_+_8992029 | 0.26 |
ENST00000543895.1
|
KLRG1
|
killer cell lectin like receptor G1 |
| chr6_-_53148822 | 0.25 |
ENST00000259803.8
|
GCM1
|
glial cells missing transcription factor 1 |
| chr7_+_95485898 | 0.24 |
ENST00000428113.5
|
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr18_-_36122110 | 0.24 |
ENST00000586829.1
|
SLC39A6
|
solute carrier family 39 member 6 |
| chr7_-_105691637 | 0.22 |
ENST00000472195.1
|
ATXN7L1
|
ataxin 7 like 1 |
| chr7_+_72925075 | 0.22 |
ENST00000434423.5
|
POM121
|
POM121 transmembrane nucleoporin |
| chr21_+_42403856 | 0.20 |
ENST00000291535.11
|
UBASH3A
|
ubiquitin associated and SH3 domain containing A |
| chr11_+_60280577 | 0.20 |
ENST00000679988.1
|
MS4A4A
|
membrane spanning 4-domains A4A |
| chr2_+_158795309 | 0.20 |
ENST00000309950.8
ENST00000621326.4 ENST00000409042.5 |
DAPL1
|
death associated protein like 1 |
| chr5_-_35047935 | 0.20 |
ENST00000510428.1
ENST00000231420.11 |
AGXT2
|
alanine--glyoxylate aminotransferase 2 |
| chr11_+_33016106 | 0.20 |
ENST00000311388.7
|
DEPDC7
|
DEP domain containing 7 |
| chr13_+_102656933 | 0.19 |
ENST00000650757.1
|
TPP2
|
tripeptidyl peptidase 2 |
| chr7_+_144003929 | 0.19 |
ENST00000408922.3
|
OR6B1
|
olfactory receptor family 6 subfamily B member 1 |
| chr8_+_97887903 | 0.18 |
ENST00000520016.5
|
MATN2
|
matrilin 2 |
| chr4_-_152411734 | 0.18 |
ENST00000603841.1
|
FBXW7
|
F-box and WD repeat domain containing 7 |
| chr12_+_53097656 | 0.18 |
ENST00000301464.4
|
IGFBP6
|
insulin like growth factor binding protein 6 |
| chr16_+_56865202 | 0.17 |
ENST00000566786.5
ENST00000438926.6 ENST00000563236.6 ENST00000262502.5 |
SLC12A3
|
solute carrier family 12 member 3 |
| chr1_-_206921867 | 0.17 |
ENST00000628511.2
ENST00000367091.8 |
FCMR
|
Fc fragment of IgM receptor |
| chr6_+_24774925 | 0.17 |
ENST00000356509.7
ENST00000230056.8 |
GMNN
|
geminin DNA replication inhibitor |
| chr18_+_22933819 | 0.15 |
ENST00000399722.6
ENST00000399725.6 ENST00000399721.6 |
RBBP8
|
RB binding protein 8, endonuclease |
| chr10_-_59362460 | 0.15 |
ENST00000422313.6
ENST00000435852.6 ENST00000614220.4 ENST00000618804.5 ENST00000621119.4 |
FAM13C
|
family with sequence similarity 13 member C |
| chr6_+_143864458 | 0.15 |
ENST00000237275.9
|
ZC2HC1B
|
zinc finger C2HC-type containing 1B |
| chr1_-_206921987 | 0.14 |
ENST00000530505.1
ENST00000442471.4 |
FCMR
|
Fc fragment of IgM receptor |
| chrX_-_71111448 | 0.14 |
ENST00000456850.6
ENST00000473378.1 ENST00000374202.7 ENST00000374188.7 |
IL2RG
|
interleukin 2 receptor subunit gamma |
| chr15_-_79971164 | 0.14 |
ENST00000335661.6
ENST00000267953.4 ENST00000677151.1 |
BCL2A1
|
BCL2 related protein A1 |
| chr3_-_197260369 | 0.14 |
ENST00000658155.1
ENST00000453607.5 |
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr17_-_3433841 | 0.14 |
ENST00000248384.1
|
OR1E2
|
olfactory receptor family 1 subfamily E member 2 |
| chr13_-_21459253 | 0.13 |
ENST00000400590.8
ENST00000382466.7 ENST00000415724.2 ENST00000542645.5 |
ZDHHC20
|
zinc finger DHHC-type palmitoyltransferase 20 |
| chr8_+_93754844 | 0.13 |
ENST00000684064.1
ENST00000498673.5 ENST00000518319.5 |
TMEM67
|
transmembrane protein 67 |
| chr21_-_30166782 | 0.13 |
ENST00000286808.5
|
CLDN17
|
claudin 17 |
| chr3_+_133124807 | 0.13 |
ENST00000508711.5
|
TMEM108
|
transmembrane protein 108 |
| chr11_+_6845683 | 0.13 |
ENST00000299454.5
|
OR10A5
|
olfactory receptor family 10 subfamily A member 5 |
| chr4_+_56505782 | 0.13 |
ENST00000640821.3
|
ARL9
|
ADP ribosylation factor like GTPase 9 |
| chr14_+_75069577 | 0.12 |
ENST00000238686.8
|
ZC2HC1C
|
zinc finger C2HC-type containing 1C |
| chr1_-_156728388 | 0.12 |
ENST00000313146.10
|
ISG20L2
|
interferon stimulated exonuclease gene 20 like 2 |
| chr16_-_71289609 | 0.12 |
ENST00000338099.9
ENST00000563876.1 |
CMTR2
|
cap methyltransferase 2 |
| chr6_+_4706133 | 0.11 |
ENST00000328908.9
|
CDYL
|
chromodomain Y like |
| chr11_+_111255982 | 0.11 |
ENST00000637637.1
|
C11orf53
|
chromosome 11 open reading frame 53 |
| chrX_-_100874351 | 0.11 |
ENST00000372966.8
|
NOX1
|
NADPH oxidase 1 |
| chr7_+_16661182 | 0.11 |
ENST00000446596.5
ENST00000452975.6 ENST00000438834.5 |
BZW2
|
basic leucine zipper and W2 domains 2 |
| chr6_-_31652678 | 0.11 |
ENST00000437771.5
ENST00000362049.10 ENST00000439687.6 ENST00000211379.9 ENST00000375964.11 ENST00000676571.1 ENST00000676615.1 |
BAG6
|
BAG cochaperone 6 |
| chrX_-_119605870 | 0.11 |
ENST00000542113.3
|
NKRF
|
NFKB repressing factor |
| chr16_-_4351283 | 0.10 |
ENST00000318059.8
|
PAM16
|
presequence translocase associated motor 16 |
| chr2_+_112721486 | 0.10 |
ENST00000327581.4
|
NT5DC4
|
5'-nucleotidase domain containing 4 |
| chr4_-_38804783 | 0.10 |
ENST00000308979.7
ENST00000505940.1 ENST00000515861.5 |
TLR1
|
toll like receptor 1 |
| chr8_+_30387064 | 0.10 |
ENST00000523115.5
ENST00000519647.5 |
RBPMS
|
RNA binding protein, mRNA processing factor |
| chr1_+_166989254 | 0.10 |
ENST00000367872.9
ENST00000447624.1 |
MAEL
|
maelstrom spermatogenic transposon silencer |
| chr5_-_83720813 | 0.10 |
ENST00000515590.1
ENST00000274341.9 |
HAPLN1
|
hyaluronan and proteoglycan link protein 1 |
| chr1_+_166989089 | 0.10 |
ENST00000367870.6
|
MAEL
|
maelstrom spermatogenic transposon silencer |
| chr5_-_95081482 | 0.10 |
ENST00000312216.12
ENST00000512425.5 ENST00000505208.5 ENST00000429576.6 ENST00000508509.5 ENST00000510732.5 |
MCTP1
|
multiple C2 and transmembrane domain containing 1 |
| chr7_+_95485934 | 0.09 |
ENST00000325885.6
|
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr5_-_72507354 | 0.09 |
ENST00000414109.2
ENST00000318442.6 |
ZNF366
|
zinc finger protein 366 |
| chr4_-_149815826 | 0.09 |
ENST00000636793.2
ENST00000636414.1 |
IQCM
|
IQ motif containing M |
| chr1_-_173603041 | 0.09 |
ENST00000367714.4
|
SLC9C2
|
solute carrier family 9 member C2 (putative) |
| chrX_-_49186328 | 0.09 |
ENST00000599218.6
ENST00000376317.4 |
PRICKLE3
|
prickle planar cell polarity protein 3 |
| chr16_+_81238682 | 0.09 |
ENST00000258168.7
ENST00000564552.1 |
BCO1
|
beta-carotene oxygenase 1 |
| chr20_+_57561103 | 0.09 |
ENST00000319441.6
|
PCK1
|
phosphoenolpyruvate carboxykinase 1 |
| chr17_+_7857695 | 0.09 |
ENST00000571846.5
|
CYB5D1
|
cytochrome b5 domain containing 1 |
| chr4_-_127965930 | 0.09 |
ENST00000641447.1
ENST00000296468.8 ENST00000641134.1 ENST00000641147.1 ENST00000641178.1 |
MFSD8
|
major facilitator superfamily domain containing 8 |
| chr3_-_194351290 | 0.09 |
ENST00000429275.1
ENST00000323830.4 |
CPN2
|
carboxypeptidase N subunit 2 |
| chr14_+_75069632 | 0.09 |
ENST00000439583.2
ENST00000554763.2 ENST00000524913.3 ENST00000525046.2 ENST00000674086.1 ENST00000526130.2 ENST00000674094.1 ENST00000532198.2 |
ZC2HC1C
|
zinc finger C2HC-type containing 1C |
| chr3_-_122383218 | 0.09 |
ENST00000479899.5
ENST00000291458.9 ENST00000497726.5 |
MIX23
|
mitochondrial matrix import factor 23 |
| chr2_+_48617798 | 0.08 |
ENST00000448460.5
ENST00000437125.5 ENST00000430487.6 |
GTF2A1L
|
general transcription factor IIA subunit 1 like |
| chr16_+_7332744 | 0.08 |
ENST00000436368.6
ENST00000311745.9 ENST00000340209.8 ENST00000620507.4 |
RBFOX1
|
RNA binding fox-1 homolog 1 |
| chr2_+_27148997 | 0.08 |
ENST00000296096.6
|
TCF23
|
transcription factor 23 |
| chr11_+_60280531 | 0.08 |
ENST00000532114.6
|
MS4A4A
|
membrane spanning 4-domains A4A |
| chrX_-_21658324 | 0.08 |
ENST00000379499.3
|
KLHL34
|
kelch like family member 34 |
| chr8_+_80485611 | 0.08 |
ENST00000379091.8
|
ZBTB10
|
zinc finger and BTB domain containing 10 |
| chr21_-_14658812 | 0.08 |
ENST00000647101.1
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr17_+_58169401 | 0.07 |
ENST00000641866.1
|
OR4D2
|
olfactory receptor family 4 subfamily D member 2 |
| chr5_+_73813518 | 0.07 |
ENST00000296799.8
|
ARHGEF28
|
Rho guanine nucleotide exchange factor 28 |
| chr17_+_7857987 | 0.07 |
ENST00000332439.5
|
CYB5D1
|
cytochrome b5 domain containing 1 |
| chr3_-_185821092 | 0.07 |
ENST00000421047.3
|
IGF2BP2
|
insulin like growth factor 2 mRNA binding protein 2 |
| chr10_+_99782628 | 0.07 |
ENST00000648689.1
ENST00000647814.1 |
ABCC2
|
ATP binding cassette subfamily C member 2 |
| chr4_-_167234266 | 0.07 |
ENST00000511269.5
ENST00000506697.5 ENST00000512042.1 |
SPOCK3
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 3 |
| chr14_-_64823148 | 0.07 |
ENST00000389722.7
|
SPTB
|
spectrin beta, erythrocytic |
| chr5_-_161546671 | 0.06 |
ENST00000517547.5
|
GABRB2
|
gamma-aminobutyric acid type A receptor subunit beta2 |
| chr21_+_42403874 | 0.06 |
ENST00000319294.11
ENST00000398367.1 |
UBASH3A
|
ubiquitin associated and SH3 domain containing A |
| chr17_+_36064265 | 0.06 |
ENST00000616054.2
|
CCL18
|
C-C motif chemokine ligand 18 |
| chr17_-_8152380 | 0.06 |
ENST00000317276.9
|
PER1
|
period circadian regulator 1 |
| chr6_-_111605859 | 0.06 |
ENST00000651359.1
ENST00000650859.1 ENST00000359831.8 ENST00000368761.11 |
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr7_-_150323489 | 0.06 |
ENST00000683684.1
ENST00000478393.5 |
ACTR3C
|
actin related protein 3C |
| chr6_-_159045104 | 0.06 |
ENST00000326965.7
|
TAGAP
|
T cell activation RhoGTPase activating protein |
| chr9_-_35812238 | 0.05 |
ENST00000396638.7
ENST00000340291.6 |
SPAG8
|
sperm associated antigen 8 |
| chr1_+_204870831 | 0.05 |
ENST00000404076.5
ENST00000539706.6 |
NFASC
|
neurofascin |
| chr7_+_69967464 | 0.05 |
ENST00000664521.1
|
AUTS2
|
activator of transcription and developmental regulator AUTS2 |
| chr1_+_15756659 | 0.05 |
ENST00000375771.5
|
FBLIM1
|
filamin binding LIM protein 1 |
| chr15_-_58279245 | 0.05 |
ENST00000558231.5
|
ALDH1A2
|
aldehyde dehydrogenase 1 family member A2 |
| chr1_+_152719522 | 0.05 |
ENST00000368775.3
|
C1orf68
|
chromosome 1 open reading frame 68 |
| chr14_-_64942783 | 0.05 |
ENST00000612794.1
|
GPX2
|
glutathione peroxidase 2 |
| chr6_+_27957241 | 0.05 |
ENST00000244623.1
|
OR2B6
|
olfactory receptor family 2 subfamily B member 6 |
| chr2_+_27628996 | 0.05 |
ENST00000616939.4
ENST00000264718.7 ENST00000610189.2 |
GPN1
|
GPN-loop GTPase 1 |
| chr17_-_40799939 | 0.05 |
ENST00000306658.8
|
KRT28
|
keratin 28 |
| chr12_-_102120065 | 0.05 |
ENST00000552283.6
ENST00000551744.2 |
NUP37
|
nucleoporin 37 |
| chr2_+_233729042 | 0.05 |
ENST00000482026.6
|
UGT1A3
|
UDP glucuronosyltransferase family 1 member A3 |
| chr1_-_157841800 | 0.05 |
ENST00000368174.5
|
CD5L
|
CD5 molecule like |
| chr8_+_24294107 | 0.05 |
ENST00000437154.6
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr8_-_81447428 | 0.04 |
ENST00000256103.3
ENST00000519260.1 |
PMP2
|
peripheral myelin protein 2 |
| chr6_-_159044980 | 0.04 |
ENST00000367066.8
|
TAGAP
|
T cell activation RhoGTPase activating protein |
| chr1_-_112618204 | 0.04 |
ENST00000369664.1
|
ST7L
|
suppression of tumorigenicity 7 like |
| chr11_+_60056653 | 0.04 |
ENST00000278865.8
|
MS4A3
|
membrane spanning 4-domains A3 |
| chr5_-_180815528 | 0.04 |
ENST00000333055.8
|
MGAT1
|
alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase |
| chr1_+_215573775 | 0.04 |
ENST00000448333.1
|
KCTD3
|
potassium channel tetramerization domain containing 3 |
| chr17_-_5419647 | 0.03 |
ENST00000573584.6
|
NUP88
|
nucleoporin 88 |
| chr1_-_173824322 | 0.03 |
ENST00000356198.6
|
CENPL
|
centromere protein L |
| chr6_-_27873525 | 0.03 |
ENST00000618305.2
|
H4C13
|
H4 clustered histone 13 |
| chr4_-_167234426 | 0.03 |
ENST00000541354.5
ENST00000509854.5 ENST00000512681.5 ENST00000357545.9 ENST00000510741.5 ENST00000510403.5 |
SPOCK3
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 3 |
| chr1_+_67207614 | 0.03 |
ENST00000425614.3
|
IL23R
|
interleukin 23 receptor |
| chr16_-_74607088 | 0.03 |
ENST00000565260.1
ENST00000447066.6 ENST00000205061.9 ENST00000422840.7 ENST00000627032.2 |
GLG1
|
golgi glycoprotein 1 |
| chr16_+_58392462 | 0.03 |
ENST00000318129.6
|
GINS3
|
GINS complex subunit 3 |
| chr7_+_107044689 | 0.03 |
ENST00000265717.5
|
PRKAR2B
|
protein kinase cAMP-dependent type II regulatory subunit beta |
| chrX_-_100874209 | 0.03 |
ENST00000372964.5
ENST00000217885.5 |
NOX1
|
NADPH oxidase 1 |
| chr14_+_23321446 | 0.03 |
ENST00000216727.9
ENST00000397276.6 |
PABPN1
|
poly(A) binding protein nuclear 1 |
| chr6_+_13272709 | 0.03 |
ENST00000379335.8
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr9_-_35079923 | 0.02 |
ENST00000378643.8
ENST00000448890.1 |
FANCG
|
FA complementation group G |
| chr8_+_7495892 | 0.02 |
ENST00000355602.3
|
DEFB107B
|
defensin beta 107B |
| chrX_-_149595314 | 0.02 |
ENST00000598963.3
|
HSFX2
|
heat shock transcription factor family, X-linked 2 |
| chr13_-_46105009 | 0.02 |
ENST00000439329.5
ENST00000674625.1 ENST00000181383.10 |
CPB2
|
carboxypeptidase B2 |
| chr8_+_24294044 | 0.02 |
ENST00000265769.9
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr5_-_161546708 | 0.02 |
ENST00000393959.6
|
GABRB2
|
gamma-aminobutyric acid type A receptor subunit beta2 |
| chr9_+_137086857 | 0.02 |
ENST00000682881.1
ENST00000683324.1 ENST00000542372.2 ENST00000683355.1 ENST00000682117.1 ENST00000682212.1 ENST00000684144.1 ENST00000683987.1 ENST00000371589.9 ENST00000535144.6 ENST00000475449.7 ENST00000684759.1 |
MAN1B1
|
mannosidase alpha class 1B member 1 |
| chrX_+_155997581 | 0.02 |
ENST00000244174.10
|
IL9R
|
interleukin 9 receptor |
| chr4_+_157220691 | 0.02 |
ENST00000509417.5
ENST00000645636.1 ENST00000296526.12 ENST00000264426.14 |
GRIA2
|
glutamate ionotropic receptor AMPA type subunit 2 |
| chrX_+_155997706 | 0.02 |
ENST00000369423.7
|
IL9R
|
interleukin 9 receptor |
| chr4_-_167234579 | 0.02 |
ENST00000502330.5
ENST00000357154.7 ENST00000421836.6 |
SPOCK3
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 3 |
| chr1_-_158554405 | 0.02 |
ENST00000641282.1
ENST00000641622.1 |
OR6Y1
|
olfactory receptor family 6 subfamily Y member 1 |
| chr13_-_23433676 | 0.02 |
ENST00000682547.1
ENST00000455470.6 ENST00000382292.9 |
SACS
|
sacsin molecular chaperone |
| chr14_-_105845677 | 0.02 |
ENST00000390556.6
|
IGHD
|
immunoglobulin heavy constant delta |
| chr1_-_149812359 | 0.02 |
ENST00000369167.2
ENST00000545683.1 |
H2BC18
|
H2B clustered histone 18 |
| chr14_-_100375333 | 0.02 |
ENST00000557297.5
ENST00000555813.5 ENST00000392882.7 ENST00000557135.5 ENST00000556698.5 ENST00000554509.5 ENST00000555410.5 |
WARS1
|
tryptophanyl-tRNA synthetase 1 |
| chr16_+_67570741 | 0.01 |
ENST00000644753.1
ENST00000642819.1 ENST00000645306.1 |
CTCF
|
CCCTC-binding factor |
| chr17_+_35844077 | 0.01 |
ENST00000604694.1
|
TAF15
|
TATA-box binding protein associated factor 15 |
| chr12_-_123272234 | 0.01 |
ENST00000544658.5
|
CDK2AP1
|
cyclin dependent kinase 2 associated protein 1 |
| chr16_+_58392391 | 0.01 |
ENST00000426538.6
ENST00000328514.11 |
GINS3
|
GINS complex subunit 3 |
| chr2_-_165204042 | 0.01 |
ENST00000283254.12
ENST00000453007.1 |
SCN3A
|
sodium voltage-gated channel alpha subunit 3 |
| chr4_+_113292925 | 0.01 |
ENST00000673353.1
ENST00000505342.6 ENST00000672915.1 ENST00000509550.5 |
ANK2
|
ankyrin 2 |
| chr6_+_29396555 | 0.01 |
ENST00000623183.1
|
OR12D2
|
olfactory receptor family 12 subfamily D member 2 |
| chr4_-_70839343 | 0.01 |
ENST00000514161.5
ENST00000499044.6 |
GRSF1
|
G-rich RNA sequence binding factor 1 |
| chr2_-_46617020 | 0.01 |
ENST00000474980.1
ENST00000281382.11 ENST00000306465.8 |
PIGF
|
phosphatidylinositol glycan anchor biosynthesis class F |
| chr7_-_83649097 | 0.01 |
ENST00000643230.2
|
SEMA3E
|
semaphorin 3E |
| chr10_-_95069489 | 0.01 |
ENST00000371270.6
ENST00000535898.5 ENST00000623108.3 |
CYP2C8
|
cytochrome P450 family 2 subfamily C member 8 |
| chr19_-_39846329 | 0.01 |
ENST00000599134.1
ENST00000597634.5 ENST00000598417.5 ENST00000601274.5 ENST00000594309.5 ENST00000221801.8 |
FBL
|
fibrillarin |
| chr2_+_170178136 | 0.01 |
ENST00000409044.7
ENST00000408978.9 |
MYO3B
|
myosin IIIB |
| chr1_+_162381703 | 0.01 |
ENST00000458626.4
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr20_-_31390483 | 0.01 |
ENST00000376315.2
|
DEFB119
|
defensin beta 119 |
| chr4_+_113292838 | 0.00 |
ENST00000672411.1
ENST00000673231.1 |
ANK2
|
ankyrin 2 |
| chr14_-_68793055 | 0.00 |
ENST00000439696.3
|
ZFP36L1
|
ZFP36 ring finger protein like 1 |
| chr2_-_70293438 | 0.00 |
ENST00000482975.6
ENST00000438261.5 |
SNRPG
|
small nuclear ribonucleoprotein polypeptide G |
| chr4_+_118685373 | 0.00 |
ENST00000388822.10
ENST00000508801.1 |
METTL14
|
methyltransferase like 14 |
| chr5_-_39364484 | 0.00 |
ENST00000263408.5
|
C9
|
complement C9 |
| chr13_-_40982880 | 0.00 |
ENST00000635415.1
|
ELF1
|
E74 like ETS transcription factor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of IRX2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0032912 | negative regulation of transforming growth factor beta2 production(GO:0032912) negative regulation of hair follicle maturation(GO:0048817) regulation of melanosome transport(GO:1902908) |
| 0.1 | 0.9 | GO:0060356 | leucine import(GO:0060356) |
| 0.1 | 0.4 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.1 | 0.4 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 | 0.8 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 | 0.3 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.0 | 0.5 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.2 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.0 | 0.1 | GO:1990451 | cellular stress response to acidic pH(GO:1990451) |
| 0.0 | 0.2 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.1 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.1 | GO:0042495 | detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.2 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.0 | 0.1 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 0.5 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:1904327 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.4 | GO:0002227 | innate immune response in mucosa(GO:0002227) positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.9 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 0.1 | GO:0016119 | carotene metabolic process(GO:0016119) |
| 0.0 | 0.1 | GO:0050787 | antibiotic metabolic process(GO:0016999) detoxification of mercury ion(GO:0050787) |
| 0.0 | 0.2 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.3 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.0 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.1 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.0 | 2.1 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.0 | 0.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.8 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.1 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0030849 | autosome(GO:0030849) |
| 0.0 | 0.9 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.1 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 0.3 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.2 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.2 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.4 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.2 | GO:0044613 | nuclear pore central transport channel(GO:0044613) nuclear pore nuclear basket(GO:0044615) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.1 | 0.9 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.8 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.4 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 1.4 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.1 | 0.8 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.9 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.2 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.0 | 0.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.2 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.0 | 0.8 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.1 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.1 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.4 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.2 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.4 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 1.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.7 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |