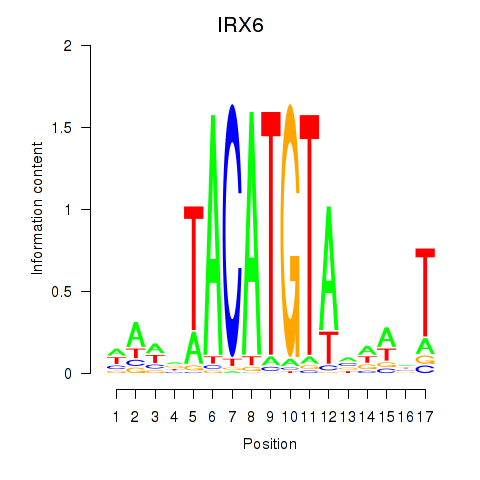
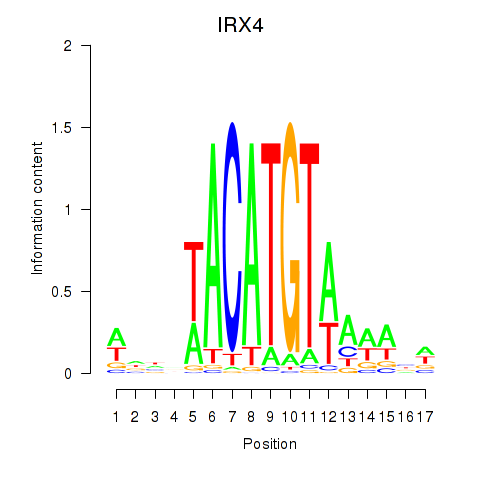
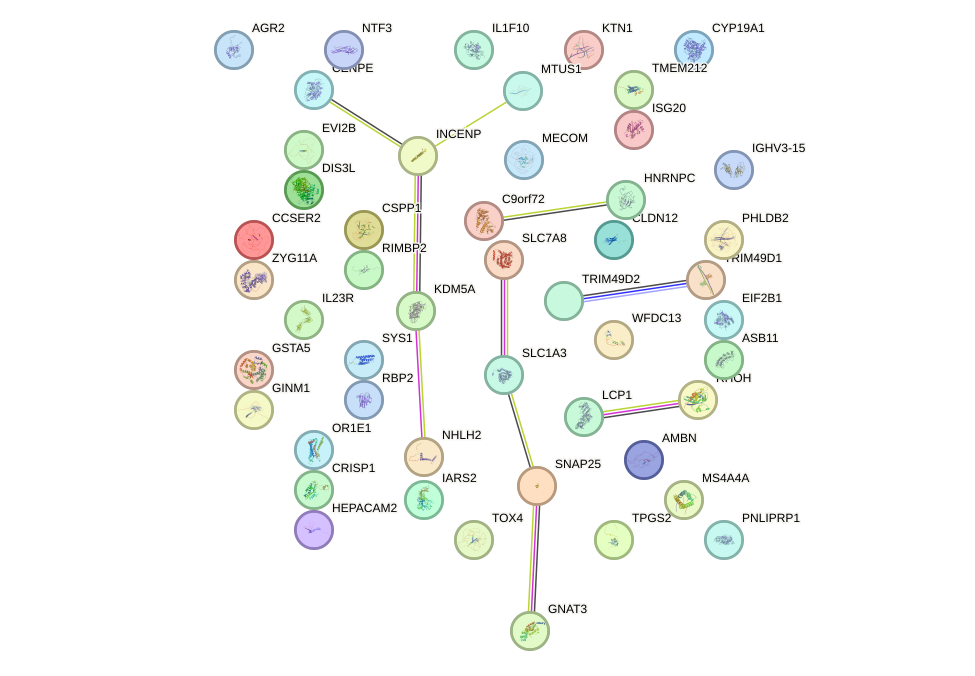
|
chr5_+_36608146
Show fit
|
0.11 |
ENST00000381918.4
ENST00000513646.1
|
SLC1A3
|
solute carrier family 1 member 3
|
|
chr3_-_27456743
Show fit
|
0.10 |
ENST00000295736.9
ENST00000428386.5
ENST00000428179.1
|
SLC4A7
|
solute carrier family 4 member 7
|
|
chr13_-_46142834
Show fit
|
0.09 |
ENST00000674665.1
|
LCP1
|
lymphocyte cytosolic protein 1
|
|
chr14_-_23154422
Show fit
|
0.09 |
ENST00000422941.6
|
SLC7A8
|
solute carrier family 7 member 8
|
|
chr14_-_23155302
Show fit
|
0.08 |
ENST00000529705.6
|
SLC7A8
|
solute carrier family 7 member 8
|
|
chr14_-_23154369
Show fit
|
0.07 |
ENST00000453702.5
|
SLC7A8
|
solute carrier family 7 member 8
|
|
chr4_+_40193642
Show fit
|
0.07 |
ENST00000617441.4
ENST00000503941.5
|
RHOH
|
ras homolog family member H
|
|
chr1_+_220094086
Show fit
|
0.06 |
ENST00000366922.3
|
IARS2
|
isoleucyl-tRNA synthetase 2, mitochondrial
|
|
chr11_+_89924064
Show fit
|
0.04 |
ENST00000623787.3
|
TRIM49D2
|
tripartite motif containing 49D2
|
|
chr21_-_32813679
Show fit
|
0.04 |
ENST00000487113.1
ENST00000382373.4
|
C21orf62
|
chromosome 21 open reading frame 62
|
|
chr4_+_70592295
Show fit
|
0.04 |
ENST00000449493.2
|
AMBN
|
ameloblastin
|
|
chr4_-_103198371
Show fit
|
0.04 |
ENST00000611174.4
ENST00000380026.8
|
CENPE
|
centromere protein E
|
|
chr21_-_32813695
Show fit
|
0.04 |
ENST00000479548.2
ENST00000490358.5
|
C21orf62
|
chromosome 21 open reading frame 62
|
|
chr17_-_31314040
Show fit
|
0.04 |
ENST00000330927.5
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr4_-_103198331
Show fit
|
0.04 |
ENST00000265148.9
ENST00000514974.1
|
CENPE
|
centromere protein E
|
|
chr7_-_16804987
Show fit
|
0.03 |
ENST00000401412.5
ENST00000419304.7
|
AGR2
|
anterior gradient 2, protein disulphide isomerase family member
|
|
chr14_+_55661272
Show fit
|
0.03 |
ENST00000555573.5
|
KTN1
|
kinectin 1
|
|
chr4_-_142560669
Show fit
|
0.03 |
ENST00000510812.5
|
INPP4B
|
inositol polyphosphate-4-phosphatase type II B
|
|
chr7_-_93226449
Show fit
|
0.03 |
ENST00000394468.7
ENST00000453812.2
|
HEPACAM2
|
HEPACAM family member 2
|
|
chr2_+_113072318
Show fit
|
0.03 |
ENST00000393197.3
|
IL1F10
|
interleukin 1 family member 10
|
|
chrY_-_6874027
Show fit
|
0.03 |
ENST00000215479.10
|
AMELY
|
amelogenin Y-linked
|
|
chr17_-_3398410
Show fit
|
0.03 |
ENST00000322608.2
|
OR1E1
|
olfactory receptor family 1 subfamily E member 1
|
|
chr11_+_62123991
Show fit
|
0.03 |
ENST00000533896.5
ENST00000278849.4
ENST00000394818.8
|
INCENP
|
inner centromere protein
|
|
chr15_+_88635626
Show fit
|
0.03 |
ENST00000379224.10
|
ISG20
|
interferon stimulated exonuclease gene 20
|
|
chr15_+_66293217
Show fit
|
0.03 |
ENST00000319194.9
ENST00000525134.6
|
DIS3L
|
DIS3 like exosome 3'-5' exoribonuclease
|
|
chr3_+_171843337
Show fit
|
0.02 |
ENST00000334567.9
ENST00000619900.4
ENST00000450693.1
|
TMEM212
|
transmembrane protein 212
|
|
chr11_+_60280577
Show fit
|
0.02 |
ENST00000679988.1
|
MS4A4A
|
membrane spanning 4-domains A4A
|
|
chr14_+_21918161
Show fit
|
0.02 |
ENST00000390439.2
|
TRAV13-2
|
T cell receptor alpha variable 13-2
|
|
chr15_+_66293541
Show fit
|
0.02 |
ENST00000319212.9
ENST00000525109.1
|
DIS3L
|
DIS3 like exosome 3'-5' exoribonuclease
|
|
chr3_-_169147734
Show fit
|
0.02 |
ENST00000464456.5
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr1_-_115841116
Show fit
|
0.02 |
ENST00000320238.3
|
NHLH2
|
nescient helix-loop-helix 2
|
|
chr12_+_5432101
Show fit
|
0.02 |
ENST00000423158.4
|
NTF3
|
neurotrophin 3
|
|
chr2_+_159733958
Show fit
|
0.02 |
ENST00000409591.5
|
MARCHF7
|
membrane associated ring-CH-type finger 7
|
|
chr14_-_106154113
Show fit
|
0.02 |
ENST00000390603.2
|
IGHV3-15
|
immunoglobulin heavy variable 3-15
|
|
chr4_-_152382522
Show fit
|
0.02 |
ENST00000296555.11
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr8_-_17697654
Show fit
|
0.02 |
ENST00000297488.10
|
MTUS1
|
microtubule associated scaffold protein 1
|
|
chr3_-_139480723
Show fit
|
0.01 |
ENST00000511956.1
ENST00000506825.1
|
RBP2
|
retinol binding protein 2
|
|
chr12_+_32502114
Show fit
|
0.01 |
ENST00000682739.1
ENST00000427716.7
ENST00000583694.2
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr14_-_21269392
Show fit
|
0.01 |
ENST00000554891.5
ENST00000555883.5
ENST00000553753.5
ENST00000555914.5
ENST00000557336.1
ENST00000555215.5
ENST00000556628.5
ENST00000555137.5
ENST00000556226.5
ENST00000555309.5
ENST00000556142.5
ENST00000554969.5
ENST00000554455.5
ENST00000556513.5
ENST00000557201.5
ENST00000420743.6
ENST00000557768.1
ENST00000553300.6
ENST00000554383.5
ENST00000554539.5
|
HNRNPC
|
heterogeneous nuclear ribonucleoprotein C
|
|
chr9_-_27573653
Show fit
|
0.01 |
ENST00000379995.1
ENST00000647196.1
ENST00000619707.5
ENST00000379997.7
|
C9orf72
|
C9orf72-SMCR8 complex subunit
|
|
chr18_-_36798482
Show fit
|
0.01 |
ENST00000590258.2
|
TPGS2
|
tubulin polyglutamylase complex subunit 2
|
|
chr2_-_39121000
Show fit
|
0.01 |
ENST00000402219.8
|
SOS1
|
SOS Ras/Rac guanine nucleotide exchange factor 1
|
|
chr10_+_116591010
Show fit
|
0.01 |
ENST00000530319.5
ENST00000527980.5
ENST00000471549.5
ENST00000534537.5
|
PNLIPRP1
|
pancreatic lipase related protein 1
|
|
chr19_+_50188180
Show fit
|
0.01 |
ENST00000598205.5
|
MYH14
|
myosin heavy chain 14
|
|
chr6_+_116369837
Show fit
|
0.01 |
ENST00000645988.1
|
DSE
|
dermatan sulfate epimerase
|
|
chr10_+_116590956
Show fit
|
0.01 |
ENST00000358834.9
ENST00000528052.5
|
PNLIPRP1
|
pancreatic lipase related protein 1
|
|
chr6_-_49866453
Show fit
|
0.01 |
ENST00000507853.5
|
CRISP1
|
cysteine rich secretory protein 1
|
|
chr6_-_49866527
Show fit
|
0.01 |
ENST00000335847.9
|
CRISP1
|
cysteine rich secretory protein 1
|
|
chr7_+_90383672
Show fit
|
0.01 |
ENST00000416322.5
|
CLDN12
|
claudin 12
|
|
chr8_+_67064316
Show fit
|
0.01 |
ENST00000675306.2
ENST00000678017.1
ENST00000262210.11
ENST00000675869.1
ENST00000677009.1
ENST00000676847.1
ENST00000676471.1
ENST00000678542.1
ENST00000677619.1
ENST00000676605.1
ENST00000678553.1
ENST00000674993.1
ENST00000678318.1
ENST00000676573.1
ENST00000676317.1
ENST00000677592.1
ENST00000679226.1
ENST00000675955.1
ENST00000676882.1
ENST00000678616.1
ENST00000678645.1
ENST00000678747.1
|
CSPP1
|
centrosome and spindle pole associated protein 1
|
|
chr11_+_60280531
Show fit
|
0.01 |
ENST00000532114.6
|
MS4A4A
|
membrane spanning 4-domains A4A
|
|
chr6_-_52840843
Show fit
|
0.01 |
ENST00000370989.6
|
GSTA5
|
glutathione S-transferase alpha 5
|
|
chr20_+_10218808
Show fit
|
0.01 |
ENST00000254976.7
ENST00000304886.6
|
SNAP25
|
synaptosome associated protein 25
|
|
chr2_+_225399684
Show fit
|
0.01 |
ENST00000636099.1
|
NYAP2
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2
|
|
chr3_+_111911604
Show fit
|
0.01 |
ENST00000495180.1
|
PHLDB2
|
pleckstrin homology like domain family B member 2
|
|
chr6_+_149566356
Show fit
|
0.00 |
ENST00000367419.10
|
GINM1
|
glycoprotein integral membrane 1
|
|
chr1_+_67207614
Show fit
|
0.00 |
ENST00000425614.3
|
IL23R
|
interleukin 23 receptor
|
|
chr7_-_80512041
Show fit
|
0.00 |
ENST00000398291.4
|
GNAT3
|
G protein subunit alpha transducin 3
|
|
chr15_-_51338575
Show fit
|
0.00 |
ENST00000557858.5
ENST00000558328.5
ENST00000396402.6
ENST00000396404.8
ENST00000561075.5
ENST00000405011.6
ENST00000559980.5
ENST00000453807.6
|
CYP19A1
|
cytochrome P450 family 19 subfamily A member 1
|
|
chr1_+_52842753
Show fit
|
0.00 |
ENST00000371528.2
|
ZYG11A
|
zyg-11 family member A, cell cycle regulator
|
|
chr10_+_84424919
Show fit
|
0.00 |
ENST00000543283.2
ENST00000494586.5
|
CCSER2
|
coiled-coil serine rich protein 2
|
|
chr20_+_45361936
Show fit
|
0.00 |
ENST00000372727.5
ENST00000414310.5
|
SYS1
|
SYS1 golgi trafficking protein
|
|
chr12_-_31792290
Show fit
|
0.00 |
ENST00000340398.5
|
H3-5
|
H3.5 histone
|
|
chr12_-_123633604
Show fit
|
0.00 |
ENST00000534960.5
ENST00000424014.7
|
EIF2B1
|
eukaryotic translation initiation factor 2B subunit alpha
|
|
chr20_+_45702030
Show fit
|
0.00 |
ENST00000305479.3
|
WFDC13
|
WAP four-disulfide core domain 13
|
|
chrX_-_15315615
Show fit
|
0.00 |
ENST00000380470.7
ENST00000480796.6
|
ASB11
|
ankyrin repeat and SOCS box containing 11
|