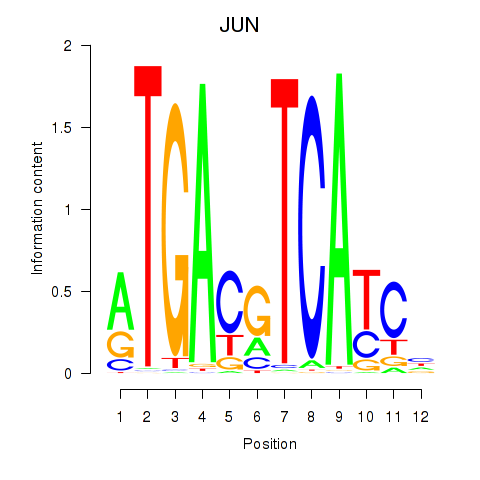
|
chrX_-_102516714
Show fit
|
0.22 |
ENST00000289373.5
|
TMSB15A
|
thymosin beta 15A
|
|
chr5_-_176537361
Show fit
|
0.21 |
ENST00000274811.9
|
RNF44
|
ring finger protein 44
|
|
chr16_-_4538819
Show fit
|
0.18 |
ENST00000564828.5
|
CDIP1
|
cell death inducing p53 target 1
|
|
chr20_+_34558706
Show fit
|
0.18 |
ENST00000360668.8
ENST00000397709.1
|
MAP1LC3A
|
microtubule associated protein 1 light chain 3 alpha
|
|
chr11_-_89490715
Show fit
|
0.17 |
ENST00000528341.5
|
NOX4
|
NADPH oxidase 4
|
|
chr5_-_172771187
Show fit
|
0.17 |
ENST00000239223.4
|
DUSP1
|
dual specificity phosphatase 1
|
|
chr2_-_237414157
Show fit
|
0.17 |
ENST00000295550.9
ENST00000353578.9
ENST00000392004.7
ENST00000433762.1
ENST00000392003.6
|
COL6A3
|
collagen type VI alpha 3 chain
|
|
chr2_-_237414127
Show fit
|
0.17 |
ENST00000472056.5
|
COL6A3
|
collagen type VI alpha 3 chain
|
|
chr14_+_67619911
Show fit
|
0.17 |
ENST00000261783.4
|
ARG2
|
arginase 2
|
|
chrX_-_93673558
Show fit
|
0.17 |
ENST00000475430.2
ENST00000373079.4
|
NAP1L3
|
nucleosome assembly protein 1 like 3
|
|
chr1_+_203305510
Show fit
|
0.16 |
ENST00000290551.5
|
BTG2
|
BTG anti-proliferation factor 2
|
|
chr6_+_89081787
Show fit
|
0.16 |
ENST00000354922.3
|
PNRC1
|
proline rich nuclear receptor coactivator 1
|
|
chr16_-_4538761
Show fit
|
0.16 |
ENST00000567695.6
ENST00000562334.5
ENST00000562579.5
ENST00000563507.5
|
CDIP1
|
cell death inducing p53 target 1
|
|
chr22_+_50090028
Show fit
|
0.15 |
ENST00000395858.7
|
MOV10L1
|
Mov10 like RISC complex RNA helicase 1
|
|
chr17_+_32991867
Show fit
|
0.15 |
ENST00000394638.1
|
SPACA3
|
sperm acrosome associated 3
|
|
chr16_-_4538469
Show fit
|
0.15 |
ENST00000588381.1
ENST00000563332.6
|
CDIP1
|
cell death inducing p53 target 1
|
|
chr1_+_1001002
Show fit
|
0.14 |
ENST00000624697.4
ENST00000624652.1
|
ISG15
|
ISG15 ubiquitin like modifier
|
|
chr9_-_96619783
Show fit
|
0.13 |
ENST00000375241.6
|
CDC14B
|
cell division cycle 14B
|
|
chr5_-_147906530
Show fit
|
0.13 |
ENST00000318315.5
ENST00000515291.1
|
C5orf46
|
chromosome 5 open reading frame 46
|
|
chr2_-_223602284
Show fit
|
0.13 |
ENST00000421386.1
ENST00000305409.3
ENST00000433889.1
|
SCG2
|
secretogranin II
|
|
chr12_+_10212867
Show fit
|
0.12 |
ENST00000545047.5
ENST00000266458.10
ENST00000629504.1
ENST00000543602.5
ENST00000545887.1
|
GABARAPL1
|
GABA type A receptor associated protein like 1
|
|
chr17_-_3298360
Show fit
|
0.12 |
ENST00000323404.2
|
OR3A1
|
olfactory receptor family 3 subfamily A member 1
|
|
chr1_-_1358524
Show fit
|
0.12 |
ENST00000445648.5
ENST00000309212.11
|
MXRA8
|
matrix remodeling associated 8
|
|
chr9_-_96619378
Show fit
|
0.12 |
ENST00000375240.7
ENST00000463569.5
|
CDC14B
|
cell division cycle 14B
|
|
chr11_-_72785932
Show fit
|
0.11 |
ENST00000539138.1
ENST00000542989.5
|
STARD10
|
StAR related lipid transfer domain containing 10
|
|
chr7_+_30134956
Show fit
|
0.10 |
ENST00000324453.13
ENST00000409688.1
|
MTURN
|
maturin, neural progenitor differentiation regulator homolog
|
|
chrX_+_87517784
Show fit
|
0.10 |
ENST00000373119.9
ENST00000373114.4
|
KLHL4
|
kelch like family member 4
|
|
chr13_-_51804094
Show fit
|
0.10 |
ENST00000280056.6
|
DHRS12
|
dehydrogenase/reductase 12
|
|
chr7_-_8262668
Show fit
|
0.10 |
ENST00000446305.1
|
ICA1
|
islet cell autoantigen 1
|
|
chr6_+_139135063
Show fit
|
0.10 |
ENST00000367658.3
|
HECA
|
hdc homolog, cell cycle regulator
|
|
chr22_+_35299800
Show fit
|
0.10 |
ENST00000456128.5
ENST00000411850.5
ENST00000425375.5
ENST00000449058.7
|
TOM1
|
target of myb1 membrane trafficking protein
|
|
chr3_-_99876104
Show fit
|
0.10 |
ENST00000471562.1
ENST00000495625.2
|
FILIP1L
|
filamin A interacting protein 1 like
|
|
chr2_-_191847068
Show fit
|
0.10 |
ENST00000304141.5
|
CAVIN2
|
caveolae associated protein 2
|
|
chr12_-_31729010
Show fit
|
0.10 |
ENST00000537562.5
ENST00000537960.5
ENST00000281471.11
ENST00000536761.5
ENST00000542781.5
ENST00000457428.6
|
AMN1
|
antagonist of mitotic exit network 1 homolog
|
|
chr5_-_111976925
Show fit
|
0.10 |
ENST00000395634.7
|
NREP
|
neuronal regeneration related protein
|
|
chr5_+_134371561
Show fit
|
0.10 |
ENST00000265339.7
ENST00000506787.5
ENST00000507277.1
|
UBE2B
|
ubiquitin conjugating enzyme E2 B
|
|
chr5_+_140564811
Show fit
|
0.09 |
ENST00000612662.2
ENST00000323146.8
ENST00000514199.1
|
SLC35A4
|
solute carrier family 35 member A4
|
|
chr9_-_137221323
Show fit
|
0.09 |
ENST00000391553.2
ENST00000392827.2
|
RNF208
|
ring finger protein 208
|
|
chr22_-_35961623
Show fit
|
0.09 |
ENST00000408983.2
|
RBFOX2
|
RNA binding fox-1 homolog 2
|
|
chr5_+_157269317
Show fit
|
0.09 |
ENST00000618329.4
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chrX_+_155380709
Show fit
|
0.09 |
ENST00000354514.6
|
H2AB2
|
H2A.B variant histone 2
|
|
chrX_+_154884972
Show fit
|
0.09 |
ENST00000620016.2
|
H2AB1
|
H2A.B variant histone 1
|
|
chr12_-_108826161
Show fit
|
0.09 |
ENST00000546697.1
|
SSH1
|
slingshot protein phosphatase 1
|
|
chr13_+_23570370
Show fit
|
0.09 |
ENST00000403372.6
ENST00000248484.9
|
TNFRSF19
|
TNF receptor superfamily member 19
|
|
chr21_-_43659460
Show fit
|
0.09 |
ENST00000443485.1
ENST00000291560.7
|
HSF2BP
|
heat shock transcription factor 2 binding protein
|
|
chr8_-_27258414
Show fit
|
0.09 |
ENST00000523048.5
|
STMN4
|
stathmin 4
|
|
chr11_-_89491131
Show fit
|
0.09 |
ENST00000343727.9
ENST00000531342.5
ENST00000375979.7
|
NOX4
|
NADPH oxidase 4
|
|
chr17_+_32991844
Show fit
|
0.08 |
ENST00000269053.8
|
SPACA3
|
sperm acrosome associated 3
|
|
chr20_-_3173516
Show fit
|
0.08 |
ENST00000360342.7
ENST00000645462.1
ENST00000337576.7
|
LZTS3
|
leucine zipper tumor suppressor family member 3
|
|
chr17_+_57256514
Show fit
|
0.08 |
ENST00000284073.7
ENST00000674964.1
|
MSI2
|
musashi RNA binding protein 2
|
|
chr11_+_64237420
Show fit
|
0.08 |
ENST00000541681.1
|
VEGFB
|
vascular endothelial growth factor B
|
|
chr9_-_34665985
Show fit
|
0.08 |
ENST00000416454.5
ENST00000544078.2
ENST00000421828.7
ENST00000423809.5
|
ENSG00000187186.15
|
novel protein
|
|
chr2_-_182242031
Show fit
|
0.08 |
ENST00000358139.6
|
PDE1A
|
phosphodiesterase 1A
|
|
chr6_+_27865308
Show fit
|
0.08 |
ENST00000613174.2
|
H2AC16
|
H2A clustered histone 16
|
|
chr11_+_6205549
Show fit
|
0.08 |
ENST00000316375.3
|
C11orf42
|
chromosome 11 open reading frame 42
|
|
chr3_+_184337591
Show fit
|
0.08 |
ENST00000383847.7
|
FAM131A
|
family with sequence similarity 131 member A
|
|
chr12_-_50396601
Show fit
|
0.08 |
ENST00000327337.6
ENST00000543111.5
|
FAM186A
|
family with sequence similarity 186 member A
|
|
chr20_-_31390580
Show fit
|
0.08 |
ENST00000339144.3
ENST00000376321.4
|
DEFB119
|
defensin beta 119
|
|
chr14_+_76761453
Show fit
|
0.08 |
ENST00000167106.9
|
VASH1
|
vasohibin 1
|
|
chr5_-_134371004
Show fit
|
0.08 |
ENST00000521755.1
ENST00000523054.5
ENST00000518409.1
|
CDKL3
ENSG00000273345.5
|
cyclin dependent kinase like 3
novel transcript
|
|
chr4_+_105552611
Show fit
|
0.08 |
ENST00000265154.6
ENST00000420470.3
|
ARHGEF38
|
Rho guanine nucleotide exchange factor 38
|
|
chr3_-_108222362
Show fit
|
0.08 |
ENST00000492106.1
|
IFT57
|
intraflagellar transport 57
|
|
chr11_-_123654939
Show fit
|
0.08 |
ENST00000657191.1
|
SCN3B
|
sodium voltage-gated channel beta subunit 3
|
|
chr3_+_159852933
Show fit
|
0.08 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr16_+_1989949
Show fit
|
0.08 |
ENST00000248121.7
ENST00000618464.1
|
SYNGR3
|
synaptogyrin 3
|
|
chr11_-_123654581
Show fit
|
0.08 |
ENST00000392770.6
ENST00000530277.5
ENST00000299333.8
|
SCN3B
|
sodium voltage-gated channel beta subunit 3
|
|
chr3_+_159273235
Show fit
|
0.08 |
ENST00000638749.1
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough
|
|
chr19_+_40588706
Show fit
|
0.08 |
ENST00000595726.1
|
SHKBP1
|
SH3KBP1 binding protein 1
|
|
chr17_-_76240478
Show fit
|
0.07 |
ENST00000269391.11
|
RNF157
|
ring finger protein 157
|
|
chr1_-_159925496
Show fit
|
0.07 |
ENST00000368097.9
|
TAGLN2
|
transgelin 2
|
|
chr1_+_77532100
Show fit
|
0.07 |
ENST00000478255.1
|
AK5
|
adenylate kinase 5
|
|
chr2_-_197310646
Show fit
|
0.07 |
ENST00000647377.1
|
ANKRD44
|
ankyrin repeat domain 44
|
|
chr19_-_50511203
Show fit
|
0.07 |
ENST00000595669.5
|
JOSD2
|
Josephin domain containing 2
|
|
chr9_+_34990250
Show fit
|
0.07 |
ENST00000454002.6
ENST00000545841.5
|
DNAJB5
|
DnaJ heat shock protein family (Hsp40) member B5
|
|
chr8_-_41665261
Show fit
|
0.07 |
ENST00000522231.5
ENST00000314214.12
ENST00000348036.8
ENST00000522543.5
|
ANK1
|
ankyrin 1
|
|
chr17_+_57256727
Show fit
|
0.07 |
ENST00000675656.1
|
MSI2
|
musashi RNA binding protein 2
|
|
chr17_-_76240289
Show fit
|
0.07 |
ENST00000647930.1
ENST00000592271.1
ENST00000319945.10
|
RNF157
|
ring finger protein 157
|
|
chr1_-_160282458
Show fit
|
0.07 |
ENST00000485079.1
|
ENSG00000258465.8
|
novel protein
|
|
chr2_-_89160329
Show fit
|
0.07 |
ENST00000390256.2
|
IGKV6-21
|
immunoglobulin kappa variable 6-21 (non-functional)
|
|
chr14_-_49586325
Show fit
|
0.07 |
ENST00000557519.1
ENST00000396020.7
ENST00000554075.2
ENST00000245458.11
|
RPS29
|
ribosomal protein S29
|
|
chr5_-_95961830
Show fit
|
0.07 |
ENST00000513343.1
ENST00000237853.9
|
ELL2
|
elongation factor for RNA polymerase II 2
|
|
chr16_-_705726
Show fit
|
0.07 |
ENST00000397621.6
ENST00000324361.9
|
FBXL16
|
F-box and leucine rich repeat protein 16
|
|
chr1_+_21981099
Show fit
|
0.07 |
ENST00000400277.2
|
CELA3B
|
chymotrypsin like elastase 3B
|
|
chr22_+_24607638
Show fit
|
0.07 |
ENST00000432867.5
|
GGT1
|
gamma-glutamyltransferase 1
|
|
chr12_-_92145838
Show fit
|
0.07 |
ENST00000256015.5
|
BTG1
|
BTG anti-proliferation factor 1
|
|
chr4_+_153152163
Show fit
|
0.07 |
ENST00000676423.1
ENST00000675745.1
ENST00000676348.1
ENST00000676408.1
ENST00000674874.1
ENST00000675315.1
ENST00000675518.1
|
TRIM2
ENSG00000288637.1
|
tripartite motif containing 2
novel protein
|
|
chr3_-_99876193
Show fit
|
0.07 |
ENST00000383694.3
|
FILIP1L
|
filamin A interacting protein 1 like
|
|
chr17_-_78903193
Show fit
|
0.07 |
ENST00000322630.3
ENST00000586713.5
|
CEP295NL
|
CEP295 N-terminal like
|
|
chr12_-_6663136
Show fit
|
0.07 |
ENST00000396807.8
ENST00000619641.4
ENST00000446105.6
|
ING4
|
inhibitor of growth family member 4
|
|
chr15_+_43791842
Show fit
|
0.07 |
ENST00000674451.1
ENST00000630046.2
|
SERF2
|
small EDRK-rich factor 2
|
|
chrX_+_111096211
Show fit
|
0.07 |
ENST00000372010.5
ENST00000519681.5
|
PAK3
|
p21 (RAC1) activated kinase 3
|
|
chr12_-_6663083
Show fit
|
0.06 |
ENST00000467678.5
ENST00000493873.1
ENST00000412586.6
ENST00000423703.6
ENST00000444704.5
ENST00000341550.9
|
ING4
|
inhibitor of growth family member 4
|
|
chr20_+_34704336
Show fit
|
0.06 |
ENST00000374809.6
ENST00000374810.8
ENST00000451665.5
|
TP53INP2
|
tumor protein p53 inducible nuclear protein 2
|
|
chr17_-_74868616
Show fit
|
0.06 |
ENST00000579893.1
ENST00000544854.5
|
FDXR
|
ferredoxin reductase
|
|
chr19_+_17933001
Show fit
|
0.06 |
ENST00000445755.7
|
CCDC124
|
coiled-coil domain containing 124
|
|
chr20_-_32536383
Show fit
|
0.06 |
ENST00000375678.7
|
NOL4L
|
nucleolar protein 4 like
|
|
chr1_-_161069666
Show fit
|
0.06 |
ENST00000368016.7
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr9_-_136764515
Show fit
|
0.06 |
ENST00000316144.6
|
LCN15
|
lipocalin 15
|
|
chr3_+_101574157
Show fit
|
0.06 |
ENST00000265260.8
ENST00000469941.5
ENST00000627393.1
|
PCNP
|
PEST proteolytic signal containing nuclear protein
|
|
chr17_-_7931910
Show fit
|
0.06 |
ENST00000303731.9
ENST00000571947.5
ENST00000540486.5
ENST00000572656.2
|
TRAPPC1
|
trafficking protein particle complex 1
|
|
chr6_+_96562548
Show fit
|
0.06 |
ENST00000541107.5
ENST00000326771.2
|
FHL5
|
four and a half LIM domains 5
|
|
chr2_+_90021567
Show fit
|
0.06 |
ENST00000436451.2
|
IGKV6D-21
|
immunoglobulin kappa variable 6D-21 (non-functional)
|
|
chr16_+_81651186
Show fit
|
0.06 |
ENST00000566462.2
|
CMIP
|
c-Maf inducing protein
|
|
chr3_-_108222383
Show fit
|
0.06 |
ENST00000264538.4
|
IFT57
|
intraflagellar transport 57
|
|
chr15_+_43792305
Show fit
|
0.06 |
ENST00000249786.9
ENST00000409960.6
ENST00000409646.5
ENST00000339624.9
ENST00000409291.5
ENST00000402131.5
ENST00000403425.5
ENST00000430901.1
|
SERF2
|
small EDRK-rich factor 2
|
|
chr5_+_141966820
Show fit
|
0.06 |
ENST00000513019.5
ENST00000394519.5
ENST00000356143.5
|
RNF14
|
ring finger protein 14
|
|
chr9_-_113299765
Show fit
|
0.06 |
ENST00000478815.1
|
RNF183
|
ring finger protein 183
|
|
chr17_+_7014745
Show fit
|
0.06 |
ENST00000546760.5
ENST00000552402.5
|
C17orf49
|
chromosome 17 open reading frame 49
|
|
chr19_-_12484773
Show fit
|
0.06 |
ENST00000397732.8
|
ZNF709
|
zinc finger protein 709
|
|
chr5_-_134174765
Show fit
|
0.06 |
ENST00000520417.1
|
SKP1
|
S-phase kinase associated protein 1
|
|
chr1_+_17205119
Show fit
|
0.06 |
ENST00000375471.5
|
PADI1
|
peptidyl arginine deiminase 1
|
|
chr1_+_29236544
Show fit
|
0.06 |
ENST00000428026.6
ENST00000460170.2
|
PTPRU
|
protein tyrosine phosphatase receptor type U
|
|
chrX_+_103962613
Show fit
|
0.06 |
ENST00000540220.6
ENST00000563257.5
ENST00000436583.5
ENST00000569577.1
|
TMSB15B
|
thymosin beta 15B
|
|
chr1_+_29236499
Show fit
|
0.06 |
ENST00000345512.7
ENST00000373779.8
|
PTPRU
|
protein tyrosine phosphatase receptor type U
|
|
chr1_-_221742074
Show fit
|
0.06 |
ENST00000366899.4
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr3_-_9952337
Show fit
|
0.06 |
ENST00000411976.2
ENST00000412055.6
|
PRRT3
|
proline rich transmembrane protein 3
|
|
chr11_-_31817904
Show fit
|
0.06 |
ENST00000423822.7
|
PAX6
|
paired box 6
|
|
chr16_+_19067639
Show fit
|
0.06 |
ENST00000568985.5
ENST00000566110.5
|
COQ7
|
coenzyme Q7, hydroxylase
|
|
chr10_+_132537814
Show fit
|
0.06 |
ENST00000368593.7
|
INPP5A
|
inositol polyphosphate-5-phosphatase A
|
|
chr2_-_197310767
Show fit
|
0.06 |
ENST00000282272.15
ENST00000409153.5
ENST00000409919.5
|
ANKRD44
|
ankyrin repeat domain 44
|
|
chr6_+_125956696
Show fit
|
0.06 |
ENST00000229633.7
|
HINT3
|
histidine triad nucleotide binding protein 3
|
|
chr3_+_185282941
Show fit
|
0.06 |
ENST00000448876.5
ENST00000446828.5
ENST00000447637.1
ENST00000424227.5
ENST00000454237.1
|
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13
|
|
chrX_+_12975083
Show fit
|
0.06 |
ENST00000451311.7
ENST00000380636.1
|
TMSB4X
|
thymosin beta 4 X-linked
|
|
chr17_+_7014774
Show fit
|
0.06 |
ENST00000439424.6
|
C17orf49
|
chromosome 17 open reading frame 49
|
|
chr1_+_85062304
Show fit
|
0.05 |
ENST00000326813.12
ENST00000528899.5
ENST00000294664.11
|
DNAI3
|
dynein axonemal intermediate chain 3
|
|
chr11_-_95232514
Show fit
|
0.05 |
ENST00000634898.1
ENST00000542176.1
ENST00000278499.6
|
SESN3
|
sestrin 3
|
|
chr16_+_19067893
Show fit
|
0.05 |
ENST00000544894.6
ENST00000561858.5
|
COQ7
|
coenzyme Q7, hydroxylase
|
|
chr4_+_174918355
Show fit
|
0.05 |
ENST00000505141.5
ENST00000359240.7
ENST00000615367.4
ENST00000445694.5
ENST00000618444.1
|
ADAM29
|
ADAM metallopeptidase domain 29
|
|
chr14_+_60249191
Show fit
|
0.05 |
ENST00000395076.9
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent 1A
|
|
chr19_-_50511173
Show fit
|
0.05 |
ENST00000598418.6
|
JOSD2
|
Josephin domain containing 2
|
|
chr18_+_13611429
Show fit
|
0.05 |
ENST00000587757.5
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4
|
|
chr11_-_66026473
Show fit
|
0.05 |
ENST00000312106.6
|
CATSPER1
|
cation channel sperm associated 1
|
|
chr10_+_122980448
Show fit
|
0.05 |
ENST00000405485.2
|
PSTK
|
phosphoseryl-tRNA kinase
|
|
chr7_-_129952901
Show fit
|
0.05 |
ENST00000472396.5
ENST00000355621.8
|
UBE2H
|
ubiquitin conjugating enzyme E2 H
|
|
chr9_+_34989641
Show fit
|
0.05 |
ENST00000453597.8
ENST00000312316.9
ENST00000458263.6
ENST00000537321.5
ENST00000682809.1
ENST00000684748.1
|
DNAJB5
|
DnaJ heat shock protein family (Hsp40) member B5
|
|
chr17_+_7015035
Show fit
|
0.05 |
ENST00000552775.1
|
C17orf49
|
chromosome 17 open reading frame 49
|
|
chr3_-_51875597
Show fit
|
0.05 |
ENST00000446461.2
|
IQCF5
|
IQ motif containing F5
|
|
chr14_-_24084625
Show fit
|
0.05 |
ENST00000397002.6
|
NRL
|
neural retina leucine zipper
|
|
chr11_+_59172116
Show fit
|
0.05 |
ENST00000227451.4
|
DTX4
|
deltex E3 ubiquitin ligase 4
|
|
chr16_+_19067989
Show fit
|
0.05 |
ENST00000569127.1
|
COQ7
|
coenzyme Q7, hydroxylase
|
|
chr3_+_126394890
Show fit
|
0.05 |
ENST00000352312.6
|
CFAP100
|
cilia and flagella associated protein 100
|
|
chrX_+_12975216
Show fit
|
0.05 |
ENST00000380635.5
|
TMSB4X
|
thymosin beta 4 X-linked
|
|
chr2_+_61162185
Show fit
|
0.05 |
ENST00000432605.3
ENST00000488469.4
|
C2orf74
|
chromosome 2 open reading frame 74
|
|
chr16_+_29962049
Show fit
|
0.05 |
ENST00000279396.11
ENST00000575829.6
ENST00000561899.6
|
TMEM219
|
transmembrane protein 219
|
|
chr19_-_55363243
Show fit
|
0.05 |
ENST00000424985.3
|
FAM71E2
|
family with sequence similarity 71 member E2
|
|
chr3_-_196942427
Show fit
|
0.05 |
ENST00000411704.1
ENST00000452404.6
|
NCBP2
|
nuclear cap binding protein subunit 2
|
|
chr11_-_2171805
Show fit
|
0.05 |
ENST00000381178.5
ENST00000381175.5
ENST00000333684.9
ENST00000352909.8
|
TH
|
tyrosine hydroxylase
|
|
chrX_+_111096136
Show fit
|
0.05 |
ENST00000372007.10
|
PAK3
|
p21 (RAC1) activated kinase 3
|
|
chr17_-_7252482
Show fit
|
0.05 |
ENST00000572043.5
|
CTDNEP1
|
CTD nuclear envelope phosphatase 1
|
|
chr19_-_50511146
Show fit
|
0.05 |
ENST00000594350.1
ENST00000601423.5
|
JOSD2
|
Josephin domain containing 2
|
|
chr19_+_57320461
Show fit
|
0.05 |
ENST00000321545.5
|
ZNF543
|
zinc finger protein 543
|
|
chr17_+_45132600
Show fit
|
0.05 |
ENST00000619916.4
ENST00000431281.5
ENST00000591859.5
|
ACBD4
|
acyl-CoA binding domain containing 4
|
|
chr15_-_51243011
Show fit
|
0.05 |
ENST00000405913.7
ENST00000559878.5
|
CYP19A1
|
cytochrome P450 family 19 subfamily A member 1
|
|
chr18_+_3262956
Show fit
|
0.05 |
ENST00000584539.1
|
MYL12B
|
myosin light chain 12B
|
|
chr21_-_34526850
Show fit
|
0.05 |
ENST00000481448.5
ENST00000381132.6
|
RCAN1
|
regulator of calcineurin 1
|
|
chr19_+_48606732
Show fit
|
0.05 |
ENST00000321762.3
|
SPACA4
|
sperm acrosome associated 4
|
|
chr6_+_27815010
Show fit
|
0.05 |
ENST00000621112.2
|
H2BC14
|
H2B clustered histone 14
|
|
chr4_-_7871986
Show fit
|
0.05 |
ENST00000360265.9
|
AFAP1
|
actin filament associated protein 1
|
|
chr5_-_132777866
Show fit
|
0.05 |
ENST00000448933.5
|
SEPTIN8
|
septin 8
|
|
chr20_+_35615812
Show fit
|
0.04 |
ENST00000679710.1
ENST00000374273.8
|
SPAG4
|
sperm associated antigen 4
|
|
chr3_-_184261547
Show fit
|
0.04 |
ENST00000296238.4
|
CAMK2N2
|
calcium/calmodulin dependent protein kinase II inhibitor 2
|
|
chr3_-_64225436
Show fit
|
0.04 |
ENST00000638394.2
|
PRICKLE2
|
prickle planar cell polarity protein 2
|
|
chr7_+_28685968
Show fit
|
0.04 |
ENST00000396298.6
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr4_-_8428424
Show fit
|
0.04 |
ENST00000514423.1
ENST00000503233.5
|
ACOX3
|
acyl-CoA oxidase 3, pristanoyl
|
|
chr1_-_204151884
Show fit
|
0.04 |
ENST00000367201.7
|
ETNK2
|
ethanolamine kinase 2
|
|
chr17_+_43025203
Show fit
|
0.04 |
ENST00000587250.4
|
RND2
|
Rho family GTPase 2
|
|
chr3_+_141738263
Show fit
|
0.04 |
ENST00000480908.1
ENST00000393000.3
ENST00000273480.4
|
RNF7
|
ring finger protein 7
|
|
chr6_-_127459364
Show fit
|
0.04 |
ENST00000487331.2
ENST00000483725.8
|
KIAA0408
|
KIAA0408
|
|
chr4_-_7872255
Show fit
|
0.04 |
ENST00000382543.4
|
AFAP1
|
actin filament associated protein 1
|
|
chr13_-_51804145
Show fit
|
0.04 |
ENST00000218981.5
ENST00000444610.8
|
DHRS12
|
dehydrogenase/reductase 12
|
|
chr5_+_69217721
Show fit
|
0.04 |
ENST00000256441.5
|
MRPS36
|
mitochondrial ribosomal protein S36
|
|
chr10_+_22345445
Show fit
|
0.04 |
ENST00000376603.6
ENST00000456231.6
ENST00000376624.8
ENST00000313311.10
ENST00000435326.5
|
SPAG6
|
sperm associated antigen 6
|
|
chr14_+_88385714
Show fit
|
0.04 |
ENST00000045347.11
|
SPATA7
|
spermatogenesis associated 7
|
|
chr1_+_27392612
Show fit
|
0.04 |
ENST00000374024.4
|
GPR3
|
G protein-coupled receptor 3
|
|
chr2_+_3658193
Show fit
|
0.04 |
ENST00000252505.4
|
ALLC
|
allantoicase
|
|
chr10_-_89207306
Show fit
|
0.04 |
ENST00000371852.4
|
CH25H
|
cholesterol 25-hydroxylase
|
|
chr11_+_117199363
Show fit
|
0.04 |
ENST00000392951.9
ENST00000525531.5
ENST00000278968.10
|
TAGLN
|
transgelin
|
|
chr11_-_14972273
Show fit
|
0.04 |
ENST00000396372.2
ENST00000361010.7
ENST00000331587.9
|
CALCA
|
calcitonin related polypeptide alpha
|
|
chr14_+_75985747
Show fit
|
0.04 |
ENST00000679083.1
ENST00000314067.11
ENST00000238628.10
ENST00000556742.1
|
IFT43
|
intraflagellar transport 43
|
|
chr2_-_144430934
Show fit
|
0.04 |
ENST00000638087.1
ENST00000638007.1
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr10_+_24239181
Show fit
|
0.04 |
ENST00000438429.5
|
KIAA1217
|
KIAA1217
|
|
chr11_-_63614425
Show fit
|
0.04 |
ENST00000415826.3
|
PLAAT3
|
phospholipase A and acyltransferase 3
|
|
chr12_-_68253502
Show fit
|
0.04 |
ENST00000328087.6
ENST00000538666.6
|
IL22
|
interleukin 22
|
|
chr13_-_100674774
Show fit
|
0.04 |
ENST00000328767.9
|
TMTC4
|
transmembrane O-mannosyltransferase targeting cadherins 4
|
|
chr9_-_122264798
Show fit
|
0.04 |
ENST00000417201.4
|
RBM18
|
RNA binding motif protein 18
|
|
chr19_-_18919348
Show fit
|
0.04 |
ENST00000349893.8
ENST00000351079.8
ENST00000600932.5
ENST00000262812.9
|
COPE
|
COPI coat complex subunit epsilon
|
|
chr12_-_120446372
Show fit
|
0.04 |
ENST00000546954.2
|
TRIAP1
|
TP53 regulated inhibitor of apoptosis 1
|
|
chr18_+_3262417
Show fit
|
0.04 |
ENST00000581193.5
ENST00000400175.9
|
MYL12B
|
myosin light chain 12B
|
|
chr19_-_8698789
Show fit
|
0.04 |
ENST00000324436.5
|
ACTL9
|
actin like 9
|
|
chr4_-_145098541
Show fit
|
0.04 |
ENST00000613466.4
ENST00000514390.5
|
ANAPC10
|
anaphase promoting complex subunit 10
|
|
chr19_+_38304105
Show fit
|
0.04 |
ENST00000588605.5
ENST00000301246.10
|
C19orf33
|
chromosome 19 open reading frame 33
|
|
chr15_-_42548763
Show fit
|
0.04 |
ENST00000563454.5
ENST00000397130.8
ENST00000570160.1
ENST00000323443.6
|
LRRC57
|
leucine rich repeat containing 57
|
|
chr11_+_111878926
Show fit
|
0.04 |
ENST00000528125.5
|
C11orf1
|
chromosome 11 open reading frame 1
|
|
chr1_+_235328959
Show fit
|
0.04 |
ENST00000643758.1
ENST00000497327.1
|
TBCE
GGPS1
|
tubulin folding cofactor E
geranylgeranyl diphosphate synthase 1
|
|
chr5_-_134226059
Show fit
|
0.04 |
ENST00000519718.1
ENST00000481195.6
|
ENSG00000272772.1
PPP2CA
|
novel protein
protein phosphatase 2 catalytic subunit alpha
|
|
chr14_+_64914361
Show fit
|
0.04 |
ENST00000607599.6
ENST00000548752.7
ENST00000551947.6
ENST00000549115.7
ENST00000552002.7
ENST00000551093.6
ENST00000549987.1
|
CHURC1
CHURC1-FNTB
|
churchill domain containing 1
CHURC1-FNTB readthrough
|
|
chr3_-_196942377
Show fit
|
0.04 |
ENST00000447325.5
|
NCBP2
|
nuclear cap binding protein subunit 2
|
|
chr3_-_182985926
Show fit
|
0.04 |
ENST00000487822.5
ENST00000460412.6
ENST00000469954.5
|
DCUN1D1
|
defective in cullin neddylation 1 domain containing 1
|
|
chr21_+_38256698
Show fit
|
0.04 |
ENST00000613499.4
ENST00000612702.4
ENST00000398925.5
ENST00000398928.5
ENST00000328656.8
ENST00000443341.5
|
KCNJ15
|
potassium inwardly rectifying channel subfamily J member 15
|
|
chr7_+_35800932
Show fit
|
0.04 |
ENST00000635172.1
ENST00000399034.7
ENST00000350320.10
ENST00000435235.6
ENST00000672279.1
ENST00000634600.1
ENST00000635047.1
|
SEPTIN7
|
septin 7
|