Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
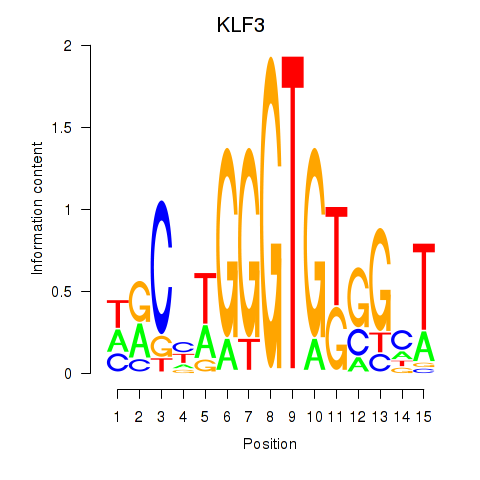
Results for KLF3
Z-value: 1.71
Transcription factors associated with KLF3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF3
|
ENSG00000109787.13 | Kruppel like factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF3 | hg38_v1_chr4_+_38664189_38664206 | 0.65 | 8.3e-02 | Click! |
Activity profile of KLF3 motif
Sorted Z-values of KLF3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_35501878 | 2.73 |
ENST00000593342.5
ENST00000601650.1 ENST00000408915.6 |
DMKN
|
dermokine |
| chr15_+_43593054 | 2.11 |
ENST00000453782.5
ENST00000300283.10 ENST00000437924.5 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr19_-_51020019 | 1.97 |
ENST00000309958.7
|
KLK10
|
kallikrein related peptidase 10 |
| chr1_+_2073462 | 1.76 |
ENST00000400921.6
|
PRKCZ
|
protein kinase C zeta |
| chr18_+_63887698 | 1.37 |
ENST00000457692.5
ENST00000299502.9 ENST00000413956.5 |
SERPINB2
|
serpin family B member 2 |
| chr20_+_46008900 | 1.32 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 |
| chr6_-_106974721 | 1.32 |
ENST00000606017.2
ENST00000620405.1 |
CD24
|
CD24 molecule |
| chr1_+_2073986 | 1.15 |
ENST00000461106.6
|
PRKCZ
|
protein kinase C zeta |
| chr6_-_136526472 | 1.12 |
ENST00000454590.5
ENST00000432797.6 |
MAP7
|
microtubule associated protein 7 |
| chr6_-_136525961 | 1.00 |
ENST00000438100.6
|
MAP7
|
microtubule associated protein 7 |
| chr11_+_59713403 | 0.99 |
ENST00000641815.1
|
STX3
|
syntaxin 3 |
| chr10_+_46375645 | 0.98 |
ENST00000622769.4
|
ANXA8L1
|
annexin A8 like 1 |
| chr10_-_47484133 | 0.97 |
ENST00000583911.5
ENST00000611843.4 |
ANXA8
|
annexin A8 |
| chr10_-_47484081 | 0.96 |
ENST00000583448.2
ENST00000583874.5 ENST00000585281.6 |
ANXA8
|
annexin A8 |
| chr19_-_51020154 | 0.96 |
ENST00000391805.5
ENST00000599077.1 |
KLK10
|
kallikrein related peptidase 10 |
| chr1_+_156061142 | 0.94 |
ENST00000361084.10
|
RAB25
|
RAB25, member RAS oncogene family |
| chr19_-_51019699 | 0.94 |
ENST00000358789.8
|
KLK10
|
kallikrein related peptidase 10 |
| chr12_-_8662073 | 0.89 |
ENST00000535411.5
ENST00000540087.5 |
MFAP5
|
microfibril associated protein 5 |
| chr16_+_67199509 | 0.88 |
ENST00000477898.5
|
ELMO3
|
engulfment and cell motility 3 |
| chr1_+_43935807 | 0.85 |
ENST00000438616.3
|
ARTN
|
artemin |
| chr11_-_5441514 | 0.81 |
ENST00000380211.1
|
OR51I1
|
olfactory receptor family 51 subfamily I member 1 |
| chr1_-_26960413 | 0.80 |
ENST00000320567.6
|
KDF1
|
keratinocyte differentiation factor 1 |
| chr7_+_121873152 | 0.78 |
ENST00000650826.1
ENST00000650728.1 ENST00000393386.7 ENST00000651390.1 ENST00000651842.1 ENST00000650681.1 |
PTPRZ1
|
protein tyrosine phosphatase receptor type Z1 |
| chr19_+_35116262 | 0.78 |
ENST00000604255.5
ENST00000344013.10 ENST00000346446.9 ENST00000603449.5 ENST00000605550.5 ENST00000604804.5 ENST00000605552.5 |
FXYD3
|
FXYD domain containing ion transport regulator 3 |
| chr1_+_34782259 | 0.77 |
ENST00000373362.3
|
GJB3
|
gap junction protein beta 3 |
| chr19_+_45340736 | 0.75 |
ENST00000391946.7
|
KLC3
|
kinesin light chain 3 |
| chr19_+_45340760 | 0.75 |
ENST00000585434.5
|
KLC3
|
kinesin light chain 3 |
| chr10_+_46375619 | 0.72 |
ENST00000584982.7
ENST00000613703.4 |
ANXA8L1
|
annexin A8 like 1 |
| chr1_-_26960369 | 0.70 |
ENST00000616918.1
|
KDF1
|
keratinocyte differentiation factor 1 |
| chr7_+_26152188 | 0.69 |
ENST00000056233.4
|
NFE2L3
|
nuclear factor, erythroid 2 like 3 |
| chr3_+_50155305 | 0.68 |
ENST00000002829.8
ENST00000426511.5 |
SEMA3F
|
semaphorin 3F |
| chr20_+_45174894 | 0.68 |
ENST00000243924.4
|
PI3
|
peptidase inhibitor 3 |
| chr1_-_183590876 | 0.67 |
ENST00000367536.5
|
NCF2
|
neutrophil cytosolic factor 2 |
| chr2_+_130836904 | 0.65 |
ENST00000409359.7
|
ARHGEF4
|
Rho guanine nucleotide exchange factor 4 |
| chr1_-_12616762 | 0.65 |
ENST00000464917.5
|
DHRS3
|
dehydrogenase/reductase 3 |
| chr12_-_85836372 | 0.64 |
ENST00000361228.5
|
RASSF9
|
Ras association domain family member 9 |
| chr3_+_111999326 | 0.63 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr13_+_36431510 | 0.62 |
ENST00000630422.2
|
CCNA1
|
cyclin A1 |
| chr1_-_209806124 | 0.61 |
ENST00000367021.8
ENST00000542854.5 |
IRF6
|
interferon regulatory factor 6 |
| chr14_-_23154422 | 0.57 |
ENST00000422941.6
|
SLC7A8
|
solute carrier family 7 member 8 |
| chr15_+_45129933 | 0.57 |
ENST00000321429.8
ENST00000389037.7 ENST00000558322.5 |
DUOX1
|
dual oxidase 1 |
| chr11_+_1839602 | 0.54 |
ENST00000617947.4
ENST00000252898.11 |
TNNI2
|
troponin I2, fast skeletal type |
| chr14_-_91867529 | 0.53 |
ENST00000435962.7
|
TC2N
|
tandem C2 domains, nuclear |
| chr17_+_75754618 | 0.53 |
ENST00000584939.1
|
ITGB4
|
integrin subunit beta 4 |
| chr15_-_72320149 | 0.53 |
ENST00000287202.10
|
CELF6
|
CUGBP Elav-like family member 6 |
| chr19_+_45340774 | 0.53 |
ENST00000589837.5
|
KLC3
|
kinesin light chain 3 |
| chr15_-_82952683 | 0.52 |
ENST00000450735.7
ENST00000304231.12 |
HOMER2
|
homer scaffold protein 2 |
| chr9_-_137054016 | 0.51 |
ENST00000312665.7
ENST00000355097.7 |
ENTPD2
|
ectonucleoside triphosphate diphosphohydrolase 2 |
| chr7_+_73830988 | 0.51 |
ENST00000340958.4
|
CLDN4
|
claudin 4 |
| chr7_+_86643902 | 0.49 |
ENST00000361669.7
|
GRM3
|
glutamate metabotropic receptor 3 |
| chr11_-_72722302 | 0.48 |
ENST00000334211.12
|
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr12_+_56080155 | 0.48 |
ENST00000267101.8
|
ERBB3
|
erb-b2 receptor tyrosine kinase 3 |
| chr14_-_99272184 | 0.48 |
ENST00000357195.8
|
BCL11B
|
BAF chromatin remodeling complex subunit BCL11B |
| chr3_+_46987972 | 0.48 |
ENST00000651747.1
|
NBEAL2
|
neurobeachin like 2 |
| chr8_+_144509049 | 0.47 |
ENST00000301327.5
|
MFSD3
|
major facilitator superfamily domain containing 3 |
| chr2_-_20225433 | 0.47 |
ENST00000381150.5
|
SDC1
|
syndecan 1 |
| chr8_+_122781621 | 0.47 |
ENST00000314393.6
|
ZHX2
|
zinc fingers and homeoboxes 2 |
| chr11_+_60924452 | 0.47 |
ENST00000453848.7
ENST00000544065.5 ENST00000005286.8 |
TMEM132A
|
transmembrane protein 132A |
| chr14_-_23154369 | 0.47 |
ENST00000453702.5
|
SLC7A8
|
solute carrier family 7 member 8 |
| chr3_+_50155024 | 0.46 |
ENST00000414301.5
ENST00000450338.5 ENST00000413852.5 |
SEMA3F
|
semaphorin 3F |
| chr1_-_28193873 | 0.46 |
ENST00000305392.3
ENST00000539896.1 |
PTAFR
|
platelet activating factor receptor |
| chr1_+_65147514 | 0.46 |
ENST00000545314.5
|
AK4
|
adenylate kinase 4 |
| chr16_+_66880503 | 0.46 |
ENST00000568869.1
ENST00000311765.4 ENST00000561704.1 ENST00000568398.1 ENST00000566776.1 |
PDP2
|
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
| chrX_-_153875847 | 0.46 |
ENST00000361699.8
ENST00000361981.7 |
L1CAM
|
L1 cell adhesion molecule |
| chr19_+_38789198 | 0.45 |
ENST00000314980.5
|
LGALS7B
|
galectin 7B |
| chr12_-_84911178 | 0.44 |
ENST00000681688.1
|
SLC6A15
|
solute carrier family 6 member 15 |
| chr6_+_79631322 | 0.44 |
ENST00000369838.6
|
SH3BGRL2
|
SH3 domain binding glutamate rich protein like 2 |
| chr11_+_1838970 | 0.44 |
ENST00000381911.6
|
TNNI2
|
troponin I2, fast skeletal type |
| chr11_-_72721908 | 0.43 |
ENST00000426523.5
ENST00000429686.5 |
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chrX_-_153886132 | 0.42 |
ENST00000370055.5
ENST00000370060.7 ENST00000420165.5 |
L1CAM
|
L1 cell adhesion molecule |
| chr19_-_51026593 | 0.42 |
ENST00000600362.5
ENST00000453757.8 ENST00000601671.1 |
KLK11
|
kallikrein related peptidase 11 |
| chr4_+_83535914 | 0.42 |
ENST00000611707.4
|
GPAT3
|
glycerol-3-phosphate acyltransferase 3 |
| chr13_+_79481124 | 0.41 |
ENST00000612570.4
ENST00000218652.11 |
NDFIP2
|
Nedd4 family interacting protein 2 |
| chr11_+_76860859 | 0.41 |
ENST00000679754.1
ENST00000534206.5 ENST00000680583.1 ENST00000532485.6 ENST00000526597.5 ENST00000533873.1 |
ACER3
|
alkaline ceramidase 3 |
| chr19_-_43670153 | 0.41 |
ENST00000601723.5
ENST00000339082.7 ENST00000340093.8 |
PLAUR
|
plasminogen activator, urokinase receptor |
| chr11_+_121576760 | 0.40 |
ENST00000532694.5
ENST00000534286.5 |
SORL1
|
sortilin related receptor 1 |
| chr19_-_11577632 | 0.40 |
ENST00000590420.1
ENST00000648477.1 |
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr3_+_142623386 | 0.39 |
ENST00000337777.7
ENST00000497199.5 |
PLS1
|
plastin 1 |
| chr3_-_171460368 | 0.39 |
ENST00000436636.7
ENST00000465393.1 ENST00000341852.10 |
TNIK
|
TRAF2 and NCK interacting kinase |
| chr1_+_13584262 | 0.39 |
ENST00000376061.8
ENST00000513143.5 |
PDPN
|
podoplanin |
| chr19_+_1067272 | 0.39 |
ENST00000590214.5
|
ARHGAP45
|
Rho GTPase activating protein 45 |
| chr20_+_62667803 | 0.39 |
ENST00000451793.1
|
SLCO4A1
|
solute carrier organic anion transporter family member 4A1 |
| chr4_+_83536097 | 0.38 |
ENST00000395226.6
ENST00000264409.5 |
GPAT3
|
glycerol-3-phosphate acyltransferase 3 |
| chr19_-_51027662 | 0.37 |
ENST00000594768.5
|
KLK11
|
kallikrein related peptidase 11 |
| chr6_+_30882914 | 0.37 |
ENST00000509639.5
ENST00000412274.6 ENST00000507901.5 ENST00000507046.5 ENST00000437124.6 ENST00000454612.6 ENST00000396342.6 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr13_+_79481468 | 0.37 |
ENST00000620924.1
|
NDFIP2
|
Nedd4 family interacting protein 2 |
| chr3_-_171460063 | 0.36 |
ENST00000284483.12
ENST00000475336.5 ENST00000357327.9 ENST00000460047.5 ENST00000488470.5 ENST00000470834.5 |
TNIK
|
TRAF2 and NCK interacting kinase |
| chr10_+_69451456 | 0.36 |
ENST00000373290.7
|
TSPAN15
|
tetraspanin 15 |
| chr20_+_62656359 | 0.35 |
ENST00000370507.5
|
SLCO4A1
|
solute carrier organic anion transporter family member 4A1 |
| chr2_+_74654228 | 0.35 |
ENST00000611975.4
ENST00000357877.7 ENST00000339773.9 ENST00000434486.5 |
SEMA4F
|
ssemaphorin 4F |
| chr13_+_79481446 | 0.34 |
ENST00000487865.5
|
NDFIP2
|
Nedd4 family interacting protein 2 |
| chr4_-_154590735 | 0.34 |
ENST00000403106.8
ENST00000622532.1 ENST00000651975.1 |
FGA
|
fibrinogen alpha chain |
| chr6_+_142301926 | 0.34 |
ENST00000296932.13
ENST00000367609.8 |
ADGRG6
|
adhesion G protein-coupled receptor G6 |
| chr10_+_46375721 | 0.33 |
ENST00000616785.1
ENST00000611655.1 ENST00000619162.5 |
ENSG00000285402.1
ANXA8L1
|
novel transcript annexin A8 like 1 |
| chr2_-_20225123 | 0.33 |
ENST00000254351.9
|
SDC1
|
syndecan 1 |
| chr9_-_83063135 | 0.32 |
ENST00000376447.4
|
RASEF
|
RAS and EF-hand domain containing |
| chr11_+_120210991 | 0.30 |
ENST00000328965.9
|
OAF
|
out at first homolog |
| chr15_-_34367159 | 0.30 |
ENST00000314891.11
|
LPCAT4
|
lysophosphatidylcholine acyltransferase 4 |
| chr6_+_142301854 | 0.30 |
ENST00000230173.10
ENST00000367608.6 |
ADGRG6
|
adhesion G protein-coupled receptor G6 |
| chr15_-_34367045 | 0.29 |
ENST00000617710.4
|
LPCAT4
|
lysophosphatidylcholine acyltransferase 4 |
| chr11_-_59212869 | 0.29 |
ENST00000361050.4
|
MPEG1
|
macrophage expressed 1 |
| chr1_-_32901330 | 0.29 |
ENST00000329151.5
ENST00000373463.8 |
TMEM54
|
transmembrane protein 54 |
| chr15_-_51751434 | 0.28 |
ENST00000558126.1
|
LYSMD2
|
LysM domain containing 2 |
| chr3_-_134029914 | 0.28 |
ENST00000493729.5
ENST00000310926.11 |
SLCO2A1
|
solute carrier organic anion transporter family member 2A1 |
| chr17_+_40015428 | 0.28 |
ENST00000394149.8
ENST00000225474.6 ENST00000331769.6 ENST00000394148.7 ENST00000577675.1 |
CSF3
|
colony stimulating factor 3 |
| chr1_+_3454657 | 0.28 |
ENST00000378378.9
|
ARHGEF16
|
Rho guanine nucleotide exchange factor 16 |
| chrX_+_153642473 | 0.27 |
ENST00000370167.8
|
DUSP9
|
dual specificity phosphatase 9 |
| chr19_+_18173804 | 0.27 |
ENST00000407280.4
|
IFI30
|
IFI30 lysosomal thiol reductase |
| chr12_+_65169546 | 0.27 |
ENST00000308330.3
|
LEMD3
|
LEM domain containing 3 |
| chr4_-_89836213 | 0.27 |
ENST00000618500.4
ENST00000508895.5 |
SNCA
|
synuclein alpha |
| chr8_+_22053543 | 0.26 |
ENST00000519850.5
ENST00000381470.7 |
DMTN
|
dematin actin binding protein |
| chr17_+_7439504 | 0.26 |
ENST00000575331.1
ENST00000293829.9 |
ENSG00000272884.1
FGF11
|
novel transcript fibroblast growth factor 11 |
| chr7_+_66205325 | 0.26 |
ENST00000304842.6
ENST00000649664.1 |
TPST1
|
tyrosylprotein sulfotransferase 1 |
| chr19_+_39196956 | 0.25 |
ENST00000339852.5
|
NCCRP1
|
NCCRP1, F-box associated domain containing |
| chr1_-_115768702 | 0.25 |
ENST00000261448.6
|
CASQ2
|
calsequestrin 2 |
| chr19_+_8053000 | 0.25 |
ENST00000390669.7
|
CCL25
|
C-C motif chemokine ligand 25 |
| chr19_-_43780957 | 0.24 |
ENST00000648319.1
|
KCNN4
|
potassium calcium-activated channel subfamily N member 4 |
| chr21_-_44928711 | 0.23 |
ENST00000517563.5
|
ITGB2
|
integrin subunit beta 2 |
| chr22_-_19854807 | 0.23 |
ENST00000416337.1
ENST00000403325.5 ENST00000453108.1 |
RTL10
GNB1L
|
retrotransposon Gag like 10 G protein subunit beta 1 like |
| chr15_+_41838839 | 0.23 |
ENST00000458483.4
|
PLA2G4B
|
phospholipase A2 group IVB |
| chr12_+_49295138 | 0.23 |
ENST00000257860.9
|
PRPH
|
peripherin |
| chr19_-_9498596 | 0.23 |
ENST00000301480.5
|
ZNF560
|
zinc finger protein 560 |
| chr18_+_58045642 | 0.23 |
ENST00000676223.1
ENST00000675147.1 |
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr1_+_206507546 | 0.22 |
ENST00000580449.5
ENST00000581503.6 |
RASSF5
|
Ras association domain family member 5 |
| chr1_-_6485433 | 0.22 |
ENST00000535355.6
|
PLEKHG5
|
pleckstrin homology and RhoGEF domain containing G5 |
| chr19_-_15898057 | 0.22 |
ENST00000011989.11
ENST00000221700.11 |
CYP4F2
|
cytochrome P450 family 4 subfamily F member 2 |
| chr15_-_51751525 | 0.22 |
ENST00000454181.6
|
LYSMD2
|
LysM domain containing 2 |
| chr2_+_29113989 | 0.22 |
ENST00000404424.5
|
CLIP4
|
CAP-Gly domain containing linker protein family member 4 |
| chr1_-_204307391 | 0.22 |
ENST00000637508.1
|
PLEKHA6
|
pleckstrin homology domain containing A6 |
| chr2_-_60550900 | 0.21 |
ENST00000643222.1
ENST00000643459.1 ENST00000489516.7 |
BCL11A
|
BAF chromatin remodeling complex subunit BCL11A |
| chr11_+_118956289 | 0.21 |
ENST00000264031.3
|
UPK2
|
uroplakin 2 |
| chr5_+_136049513 | 0.21 |
ENST00000514554.5
|
TGFBI
|
transforming growth factor beta induced |
| chr15_-_75712828 | 0.21 |
ENST00000308508.5
|
CSPG4
|
chondroitin sulfate proteoglycan 4 |
| chr19_+_8052315 | 0.20 |
ENST00000680646.1
|
CCL25
|
C-C motif chemokine ligand 25 |
| chr1_+_15756659 | 0.20 |
ENST00000375771.5
|
FBLIM1
|
filamin binding LIM protein 1 |
| chr19_+_8052335 | 0.20 |
ENST00000680507.1
ENST00000680450.1 ENST00000681526.1 ENST00000680506.1 |
CCL25
|
C-C motif chemokine ligand 25 |
| chr19_+_8209300 | 0.20 |
ENST00000558268.5
ENST00000558331.5 |
CERS4
|
ceramide synthase 4 |
| chr2_-_135985568 | 0.20 |
ENST00000264161.9
|
DARS1
|
aspartyl-tRNA synthetase 1 |
| chr2_+_118088432 | 0.20 |
ENST00000245787.9
|
INSIG2
|
insulin induced gene 2 |
| chr14_+_77181780 | 0.20 |
ENST00000298351.5
ENST00000554346.5 |
TMEM63C
|
transmembrane protein 63C |
| chr13_-_98977975 | 0.20 |
ENST00000376460.5
|
DOCK9
|
dedicator of cytokinesis 9 |
| chr8_-_33567118 | 0.20 |
ENST00000256257.2
|
RNF122
|
ring finger protein 122 |
| chr17_-_64130125 | 0.19 |
ENST00000680433.1
ENST00000433197.4 |
ERN1
|
endoplasmic reticulum to nucleus signaling 1 |
| chr7_+_74209386 | 0.19 |
ENST00000344995.9
|
LAT2
|
linker for activation of T cells family member 2 |
| chr17_+_16217198 | 0.19 |
ENST00000581006.5
ENST00000584797.5 ENST00000225609.10 ENST00000395844.8 ENST00000463810.2 |
PIGL
|
phosphatidylinositol glycan anchor biosynthesis class L |
| chr8_-_143617498 | 0.19 |
ENST00000425753.7
|
GFUS
|
GDP-L-fucose synthase |
| chr14_-_21023318 | 0.19 |
ENST00000298684.9
ENST00000557169.5 ENST00000553563.5 |
NDRG2
|
NDRG family member 2 |
| chr11_+_77179750 | 0.19 |
ENST00000458169.2
|
MYO7A
|
myosin VIIA |
| chrX_-_153944621 | 0.18 |
ENST00000393700.8
|
RENBP
|
renin binding protein |
| chr18_-_63661884 | 0.18 |
ENST00000332821.8
ENST00000283752.10 |
SERPINB3
|
serpin family B member 3 |
| chr3_-_195892706 | 0.18 |
ENST00000673038.1
ENST00000678220.1 ENST00000428187.7 |
TNK2
|
tyrosine kinase non receptor 2 |
| chr15_-_42273454 | 0.18 |
ENST00000448392.5
|
TMEM87A
|
transmembrane protein 87A |
| chr19_+_8052752 | 0.18 |
ENST00000315626.6
ENST00000253451.9 |
CCL25
|
C-C motif chemokine ligand 25 |
| chr21_-_46142701 | 0.18 |
ENST00000494498.2
|
FTCD
|
formimidoyltransferase cyclodeaminase |
| chr1_+_207321668 | 0.18 |
ENST00000367064.9
ENST00000314754.12 ENST00000367067.8 ENST00000644836.1 ENST00000343420.6 |
CD55
|
CD55 molecule (Cromer blood group) |
| chr19_+_8209320 | 0.18 |
ENST00000561053.5
ENST00000559450.5 ENST00000251363.10 ENST00000559336.5 |
CERS4
|
ceramide synthase 4 |
| chr21_-_44262216 | 0.17 |
ENST00000270172.7
|
DNMT3L
|
DNA methyltransferase 3 like |
| chrX_-_153944655 | 0.17 |
ENST00000369997.7
|
RENBP
|
renin binding protein |
| chr18_+_44697118 | 0.17 |
ENST00000677077.1
|
SETBP1
|
SET binding protein 1 |
| chr8_+_144358974 | 0.17 |
ENST00000526891.2
|
SLC52A2
|
solute carrier family 52 member 2 |
| chr22_+_20967212 | 0.17 |
ENST00000434714.6
|
AIFM3
|
apoptosis inducing factor mitochondria associated 3 |
| chr17_-_75289212 | 0.17 |
ENST00000582778.1
ENST00000581988.5 ENST00000579207.5 ENST00000583332.5 ENST00000442286.6 ENST00000580151.5 ENST00000580994.5 ENST00000584438.1 ENST00000416858.7 ENST00000320362.7 |
SLC25A19
|
solute carrier family 25 member 19 |
| chr12_-_214146 | 0.17 |
ENST00000684302.1
|
SLC6A12
|
solute carrier family 6 member 12 |
| chr10_-_112183698 | 0.17 |
ENST00000369425.5
ENST00000348367.9 |
GPAM
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr22_+_20967243 | 0.17 |
ENST00000683034.1
ENST00000440238.4 ENST00000405089.5 |
AIFM3
|
apoptosis inducing factor mitochondria associated 3 |
| chr12_-_101830799 | 0.17 |
ENST00000549940.5
ENST00000392919.4 |
GNPTAB
|
N-acetylglucosamine-1-phosphate transferase subunits alpha and beta |
| chr12_-_213656 | 0.17 |
ENST00000359674.8
|
SLC6A12
|
solute carrier family 6 member 12 |
| chr5_+_148063971 | 0.17 |
ENST00000398454.5
ENST00000359874.7 ENST00000508733.5 ENST00000256084.8 |
SPINK5
|
serine peptidase inhibitor Kazal type 5 |
| chr11_-_114595777 | 0.17 |
ENST00000375478.4
|
NXPE4
|
neurexophilin and PC-esterase domain family member 4 |
| chr11_+_1410443 | 0.17 |
ENST00000528710.5
|
BRSK2
|
BR serine/threonine kinase 2 |
| chr10_-_42638551 | 0.17 |
ENST00000493285.2
ENST00000359467.8 |
ZNF33B
|
zinc finger protein 33B |
| chr11_-_114595750 | 0.16 |
ENST00000424261.6
|
NXPE4
|
neurexophilin and PC-esterase domain family member 4 |
| chr3_-_130746760 | 0.16 |
ENST00000356763.8
|
PIK3R4
|
phosphoinositide-3-kinase regulatory subunit 4 |
| chr11_+_6205549 | 0.16 |
ENST00000316375.3
|
C11orf42
|
chromosome 11 open reading frame 42 |
| chr1_+_207321532 | 0.16 |
ENST00000367063.6
ENST00000391921.9 ENST00000645323.1 |
CD55
|
CD55 molecule (Cromer blood group) |
| chr8_-_143617457 | 0.16 |
ENST00000529048.5
ENST00000529064.5 |
GFUS
|
GDP-L-fucose synthase |
| chr16_+_46689640 | 0.16 |
ENST00000219097.7
ENST00000568364.6 |
ORC6
|
origin recognition complex subunit 6 |
| chr17_-_29761390 | 0.16 |
ENST00000324677.12
|
SSH2
|
slingshot protein phosphatase 2 |
| chr6_+_37257762 | 0.16 |
ENST00000373491.3
|
TBC1D22B
|
TBC1 domain family member 22B |
| chr13_-_46438190 | 0.16 |
ENST00000409879.6
|
RUBCNL
|
rubicon like autophagy enhancer |
| chr3_-_14124816 | 0.16 |
ENST00000295767.9
ENST00000396914.4 |
CHCHD4
|
coiled-coil-helix-coiled-coil-helix domain containing 4 |
| chr2_-_25982471 | 0.16 |
ENST00000264712.8
|
KIF3C
|
kinesin family member 3C |
| chr15_-_42273408 | 0.16 |
ENST00000389834.9
ENST00000307216.10 |
TMEM87A
|
transmembrane protein 87A |
| chr6_-_132714045 | 0.15 |
ENST00000367928.5
|
VNN1
|
vanin 1 |
| chr15_-_62165274 | 0.15 |
ENST00000380392.4
|
C2CD4B
|
C2 calcium dependent domain containing 4B |
| chr6_-_20212403 | 0.15 |
ENST00000324607.8
|
MBOAT1
|
membrane bound O-acyltransferase domain containing 1 |
| chr17_+_49132763 | 0.15 |
ENST00000393354.7
|
B4GALNT2
|
beta-1,4-N-acetyl-galactosaminyltransferase 2 |
| chr8_-_10655137 | 0.15 |
ENST00000382483.4
|
RP1L1
|
RP1 like 1 |
| chr1_+_99970430 | 0.15 |
ENST00000370153.6
|
SLC35A3
|
solute carrier family 35 member A3 |
| chr14_+_103100328 | 0.15 |
ENST00000559116.1
|
EXOC3L4
|
exocyst complex component 3 like 4 |
| chr19_+_37371152 | 0.15 |
ENST00000483919.5
ENST00000588911.5 ENST00000587349.1 |
ZNF527
|
zinc finger protein 527 |
| chr2_-_55917699 | 0.15 |
ENST00000634374.1
|
EFEMP1
|
EGF containing fibulin extracellular matrix protein 1 |
| chr19_+_15640880 | 0.15 |
ENST00000586182.6
ENST00000221307.13 ENST00000591058.5 |
CYP4F3
|
cytochrome P450 family 4 subfamily F member 3 |
| chr12_+_52010485 | 0.15 |
ENST00000552049.5
ENST00000546756.1 |
TAMALIN
|
trafficking regulator and scaffold protein tamalin |
| chr22_+_31082860 | 0.15 |
ENST00000619644.4
|
SMTN
|
smoothelin |
| chr10_-_97401277 | 0.15 |
ENST00000315563.10
ENST00000370992.9 ENST00000414986.5 |
RRP12
|
ribosomal RNA processing 12 homolog |
| chr17_-_39166138 | 0.15 |
ENST00000269586.12
|
ARL5C
|
ADP ribosylation factor like GTPase 5C |
| chr19_-_43781249 | 0.14 |
ENST00000615047.4
|
KCNN4
|
potassium calcium-activated channel subfamily N member 4 |
| chr14_-_24299707 | 0.14 |
ENST00000288111.12
|
DHRS1
|
dehydrogenase/reductase 1 |
| chr12_+_48122574 | 0.14 |
ENST00000549022.5
ENST00000547587.5 ENST00000312352.11 |
PFKM
|
phosphofructokinase, muscle |
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.7 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.5 | 2.9 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.5 | 1.9 | GO:1900138 | negative regulation of phospholipase A2 activity(GO:1900138) |
| 0.3 | 1.3 | GO:0032595 | B cell receptor transport within lipid bilayer(GO:0032595) B cell receptor transport into membrane raft(GO:0032597) protein transport out of membrane raft(GO:0032599) chemokine receptor transport out of membrane raft(GO:0032600) negative regulation of transforming growth factor beta3 production(GO:0032913) chemokine receptor transport within lipid bilayer(GO:0033606) |
| 0.2 | 0.9 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.2 | 1.7 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.2 | 1.5 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.2 | 0.8 | GO:0048627 | myoblast development(GO:0048627) |
| 0.2 | 0.9 | GO:0097021 | Peyer's patch morphogenesis(GO:0061146) lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.2 | 1.1 | GO:0036484 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.1 | 0.6 | GO:0042335 | cuticle development(GO:0042335) |
| 0.1 | 0.4 | GO:1902771 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.1 | 0.4 | GO:1904328 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.1 | 0.4 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.1 | 0.4 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 0.2 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.1 | 0.8 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 | 1.0 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 0.1 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) |
| 0.1 | 0.3 | GO:1990569 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.1 | 0.5 | GO:1902943 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) regulation of voltage-gated chloride channel activity(GO:1902941) positive regulation of voltage-gated chloride channel activity(GO:1902943) regulation of leukocyte tethering or rolling(GO:1903236) positive regulation of leukocyte tethering or rolling(GO:1903238) |
| 0.1 | 0.4 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 0.4 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 0.4 | GO:0009181 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.1 | 0.8 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 0.3 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.1 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.1 | 0.2 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.1 | 0.3 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 0.2 | GO:0030974 | thiamine pyrophosphate transport(GO:0030974) |
| 0.1 | 0.7 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.2 | GO:1904800 | negative regulation of dendrite extension(GO:1903860) regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.1 | 0.4 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.2 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.6 | GO:0010578 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.0 | 0.8 | GO:0070444 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.2 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.0 | 0.1 | GO:0072334 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.0 | 0.1 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.5 | GO:0036289 | peptidyl-serine autophosphorylation(GO:0036289) |
| 0.0 | 0.3 | GO:0051620 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.0 | 0.2 | GO:2000213 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.0 | 0.5 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 1.0 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.3 | GO:1901731 | positive regulation of platelet aggregation(GO:1901731) |
| 0.0 | 0.0 | GO:0038129 | ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 1.5 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.9 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.0 | 0.2 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.0 | 0.2 | GO:1904021 | negative regulation of G-protein coupled receptor internalization(GO:1904021) |
| 0.0 | 0.6 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.9 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.1 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 0.4 | GO:0048563 | post-embryonic organ morphogenesis(GO:0048563) |
| 0.0 | 0.5 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.2 | GO:1901523 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.0 | 0.5 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.1 | GO:0060458 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) positive regulation of immature T cell proliferation in thymus(GO:0033092) right lung development(GO:0060458) primary prostatic bud elongation(GO:0060516) epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.0 | 0.5 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.4 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.1 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 | 0.1 | GO:1901877 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.0 | 1.0 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.5 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.1 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.0 | 0.1 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.1 | GO:0006726 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.0 | 0.2 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.0 | 0.3 | GO:0086068 | Purkinje myocyte to ventricular cardiac muscle cell signaling(GO:0086029) Purkinje myocyte to ventricular cardiac muscle cell communication(GO:0086068) |
| 0.0 | 0.3 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.4 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.0 | 0.1 | GO:1904761 | negative regulation of myofibroblast differentiation(GO:1904761) |
| 0.0 | 0.5 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.2 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.6 | GO:0007620 | copulation(GO:0007620) |
| 0.0 | 0.3 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.5 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 0.4 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.6 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 | 0.2 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.8 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 1.8 | GO:0008088 | axo-dendritic transport(GO:0008088) |
| 0.0 | 0.1 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 | 0.1 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.7 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.3 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.1 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.8 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.1 | GO:0036245 | cellular response to menadione(GO:0036245) |
| 0.0 | 0.1 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.0 | 2.2 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.0 | 0.1 | GO:0001189 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.0 | 0.1 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.3 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.0 | 0.9 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.1 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0006258 | UDP-glucose catabolic process(GO:0006258) negative regulation of type B pancreatic cell development(GO:2000077) |
| 0.0 | 0.5 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.2 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.0 | 0.2 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 | 0.1 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.1 | GO:0060681 | branch elongation involved in ureteric bud branching(GO:0060681) |
| 0.0 | 1.0 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.2 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.0 | 0.2 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.0 | 0.1 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.2 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.0 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 0.0 | 0.2 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.0 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 0.2 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 1.0 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.0 | 0.3 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.0 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 0.0 | 0.4 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 0.2 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.0 | GO:0043203 | axon hillock(GO:0043203) |
| 0.2 | 1.0 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.2 | 0.8 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.2 | 0.6 | GO:0097123 | cyclin A1-CDK2 complex(GO:0097123) |
| 0.1 | 2.0 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 0.4 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.7 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.1 | 0.7 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 0.3 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 0.2 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 1.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 1.0 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.0 | 1.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.9 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.7 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.2 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.0 | 0.5 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.6 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.8 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.4 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.7 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.2 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.2 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.0 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.3 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.3 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.4 | GO:0005605 | basal lamina(GO:0005605) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.2 | 1.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.6 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.1 | 0.8 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.1 | 0.4 | GO:0097258 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.1 | 0.4 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 2.9 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.1 | 0.5 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.1 | 0.5 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 0.3 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.1 | 0.7 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 0.5 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.1 | 1.0 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 0.2 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.1 | 0.4 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.1 | 1.0 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.1 | 1.0 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 1.0 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 0.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 1.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.5 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.2 | GO:0090422 | thiamine pyrophosphate transporter activity(GO:0090422) |
| 0.1 | 0.7 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.2 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 0.1 | 1.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.9 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 0.4 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.7 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.0 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.6 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.6 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.3 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.2 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.4 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.8 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.2 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.2 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.8 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:1901474 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 1.3 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.9 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0003974 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.0 | 2.0 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.3 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.5 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.2 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.0 | 0.4 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.2 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.1 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.4 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.8 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0001626 | nociceptin receptor activity(GO:0001626) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 0.4 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.1 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.2 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 1.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.3 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 5.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.1 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.0 | 0.0 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.4 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.3 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.1 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.5 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.0 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.0 | 1.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.1 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.0 | 0.4 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.1 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.1 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.0 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.1 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.9 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.6 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.7 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.4 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 3.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.5 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.6 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 2.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.7 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.1 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.0 | 1.0 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.8 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 1.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.8 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.5 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.6 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.6 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.2 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |