Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
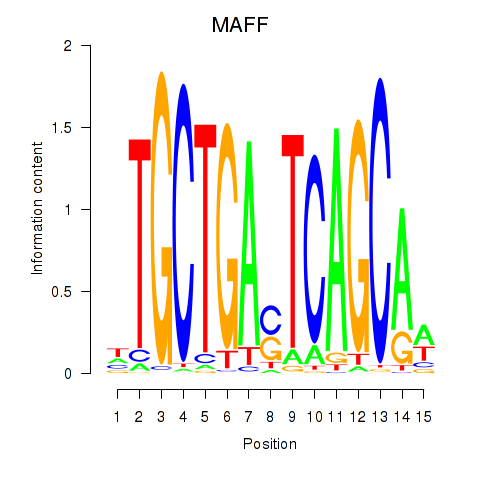
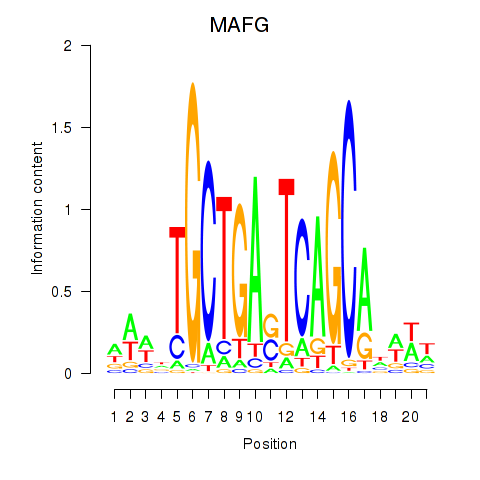
Results for MAFF_MAFG
Z-value: 0.80
Transcription factors associated with MAFF_MAFG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MAFF
|
ENSG00000185022.12 | MAF bZIP transcription factor F |
|
MAFG
|
ENSG00000197063.11 | MAF bZIP transcription factor G |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MAFF | hg38_v1_chr22_+_38213530_38213541, hg38_v1_chr22_+_38201932_38202014 | 0.86 | 6.0e-03 | Click! |
| MAFG | hg38_v1_chr17_-_81923532_81923547 | 0.83 | 1.1e-02 | Click! |
Activity profile of MAFF_MAFG motif
Sorted Z-values of MAFF_MAFG motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_190122317 | 0.66 |
ENST00000427335.6
|
P3H2
|
prolyl 3-hydroxylase 2 |
| chr12_-_84911178 | 0.65 |
ENST00000681688.1
|
SLC6A15
|
solute carrier family 6 member 15 |
| chr19_+_35118456 | 0.65 |
ENST00000604621.5
|
FXYD3
|
FXYD domain containing ion transport regulator 3 |
| chr16_+_4788411 | 0.64 |
ENST00000589327.5
|
SMIM22
|
small integral membrane protein 22 |
| chr16_+_4788394 | 0.63 |
ENST00000615471.4
ENST00000589721.5 ENST00000615889.4 |
SMIM22
|
small integral membrane protein 22 |
| chr1_-_62319264 | 0.60 |
ENST00000354381.3
|
KANK4
|
KN motif and ankyrin repeat domains 4 |
| chr1_-_62319425 | 0.56 |
ENST00000371153.9
|
KANK4
|
KN motif and ankyrin repeat domains 4 |
| chr1_+_89364051 | 0.55 |
ENST00000370456.5
|
GBP6
|
guanylate binding protein family member 6 |
| chr4_-_170003738 | 0.55 |
ENST00000502832.1
ENST00000393704.3 |
MFAP3L
|
microfibril associated protein 3 like |
| chr1_+_153031195 | 0.54 |
ENST00000307098.5
|
SPRR1B
|
small proline rich protein 1B |
| chr20_+_62656359 | 0.54 |
ENST00000370507.5
|
SLCO4A1
|
solute carrier organic anion transporter family member 4A1 |
| chr1_-_153460644 | 0.53 |
ENST00000368723.4
|
S100A7
|
S100 calcium binding protein A7 |
| chr7_-_22194709 | 0.52 |
ENST00000458533.5
|
RAPGEF5
|
Rap guanine nucleotide exchange factor 5 |
| chr1_+_2073986 | 0.45 |
ENST00000461106.6
|
PRKCZ
|
protein kinase C zeta |
| chrX_+_139530730 | 0.45 |
ENST00000218099.7
|
F9
|
coagulation factor IX |
| chr1_+_2073462 | 0.41 |
ENST00000400921.6
|
PRKCZ
|
protein kinase C zeta |
| chr17_-_8119047 | 0.39 |
ENST00000318227.4
|
ALOXE3
|
arachidonate lipoxygenase 3 |
| chr18_+_63775395 | 0.36 |
ENST00000398019.7
|
SERPINB7
|
serpin family B member 7 |
| chr5_-_1886938 | 0.35 |
ENST00000613726.4
|
IRX4
|
iroquois homeobox 4 |
| chr11_-_102798148 | 0.35 |
ENST00000315274.7
|
MMP1
|
matrix metallopeptidase 1 |
| chr17_-_41060109 | 0.34 |
ENST00000391418.3
|
KRTAP2-3
|
keratin associated protein 2-3 |
| chr6_-_47042306 | 0.34 |
ENST00000371253.7
|
ADGRF1
|
adhesion G protein-coupled receptor F1 |
| chr5_+_167754918 | 0.34 |
ENST00000519204.5
|
TENM2
|
teneurin transmembrane protein 2 |
| chr2_+_102337148 | 0.32 |
ENST00000311734.6
ENST00000409584.5 |
IL1RL1
|
interleukin 1 receptor like 1 |
| chr2_+_197705353 | 0.31 |
ENST00000282276.8
|
MARS2
|
methionyl-tRNA synthetase 2, mitochondrial |
| chr2_-_31138041 | 0.31 |
ENST00000324589.9
|
GALNT14
|
polypeptide N-acetylgalactosaminyltransferase 14 |
| chr12_+_20815672 | 0.31 |
ENST00000261196.6
ENST00000381541.7 ENST00000540229.1 |
SLCO1B3
SLCO1B3-SLCO1B7
|
solute carrier organic anion transporter family member 1B3 SLCO1B3-SLCO1B7 readthrough |
| chr10_-_114684612 | 0.29 |
ENST00000533213.6
ENST00000369252.8 |
ABLIM1
|
actin binding LIM protein 1 |
| chr2_+_102008515 | 0.28 |
ENST00000441002.1
|
IL1R2
|
interleukin 1 receptor type 2 |
| chr10_-_114684457 | 0.28 |
ENST00000392955.7
|
ABLIM1
|
actin binding LIM protein 1 |
| chr6_-_47309898 | 0.26 |
ENST00000296861.2
|
TNFRSF21
|
TNF receptor superfamily member 21 |
| chr11_+_60280577 | 0.26 |
ENST00000679988.1
|
MS4A4A
|
membrane spanning 4-domains A4A |
| chr9_-_5833014 | 0.26 |
ENST00000339450.10
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1 |
| chr9_-_137054016 | 0.25 |
ENST00000312665.7
ENST00000355097.7 |
ENTPD2
|
ectonucleoside triphosphate diphosphohydrolase 2 |
| chr11_+_1834415 | 0.23 |
ENST00000381968.7
ENST00000381978.7 |
SYT8
|
synaptotagmin 8 |
| chr1_+_206583255 | 0.23 |
ENST00000581888.1
|
RASSF5
|
Ras association domain family member 5 |
| chr17_+_7440738 | 0.23 |
ENST00000575398.5
ENST00000575082.5 |
FGF11
|
fibroblast growth factor 11 |
| chr16_+_82035245 | 0.23 |
ENST00000199936.9
|
HSD17B2
|
hydroxysteroid 17-beta dehydrogenase 2 |
| chr11_+_60280658 | 0.23 |
ENST00000337908.5
|
MS4A4A
|
membrane spanning 4-domains A4A |
| chr12_-_10674013 | 0.23 |
ENST00000535345.5
ENST00000542562.5 ENST00000075503.8 |
STYK1
|
serine/threonine/tyrosine kinase 1 |
| chr5_+_100535317 | 0.22 |
ENST00000312637.5
|
FAM174A
|
family with sequence similarity 174 member A |
| chr17_-_3964291 | 0.22 |
ENST00000359983.7
ENST00000352011.7 |
ATP2A3
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 3 |
| chr1_-_153057504 | 0.22 |
ENST00000392653.3
|
SPRR2A
|
small proline rich protein 2A |
| chr5_-_176630364 | 0.22 |
ENST00000310112.7
|
SNCB
|
synuclein beta |
| chr14_-_23155302 | 0.21 |
ENST00000529705.6
|
SLC7A8
|
solute carrier family 7 member 8 |
| chr19_+_38930916 | 0.21 |
ENST00000308018.9
ENST00000407800.2 ENST00000402029.3 |
MRPS12
|
mitochondrial ribosomal protein S12 |
| chr6_-_138107412 | 0.21 |
ENST00000421351.4
|
PERP
|
p53 apoptosis effector related to PMP22 |
| chr5_-_102498913 | 0.21 |
ENST00000513675.1
ENST00000379807.7 ENST00000506729.6 |
SLCO6A1
|
solute carrier organic anion transporter family member 6A1 |
| chr1_-_110070654 | 0.20 |
ENST00000647563.2
|
ALX3
|
ALX homeobox 3 |
| chr2_+_119431846 | 0.20 |
ENST00000306406.5
|
TMEM37
|
transmembrane protein 37 |
| chr5_-_176630517 | 0.19 |
ENST00000393693.7
ENST00000614675.4 |
SNCB
|
synuclein beta |
| chr3_+_143001562 | 0.19 |
ENST00000473835.7
ENST00000493598.6 |
U2SURP
|
U2 snRNP associated SURP domain containing |
| chr10_+_24208774 | 0.18 |
ENST00000376456.8
ENST00000458595.5 ENST00000376452.7 ENST00000430453.6 |
KIAA1217
|
KIAA1217 |
| chr5_-_102499008 | 0.18 |
ENST00000389019.7
|
SLCO6A1
|
solute carrier organic anion transporter family member 6A1 |
| chr6_+_106098933 | 0.18 |
ENST00000369089.3
|
PRDM1
|
PR/SET domain 1 |
| chr17_+_8039034 | 0.18 |
ENST00000572022.5
ENST00000380173.6 ENST00000380183.9 |
ALOX15B
|
arachidonate 15-lipoxygenase type B |
| chr7_-_22500152 | 0.17 |
ENST00000406890.6
ENST00000678116.1 ENST00000424363.5 |
STEAP1B
|
STEAP family member 1B |
| chr18_-_55401751 | 0.17 |
ENST00000537856.7
|
TCF4
|
transcription factor 4 |
| chr9_-_133149334 | 0.17 |
ENST00000393160.7
|
RALGDS
|
ral guanine nucleotide dissociation stimulator |
| chr13_+_24270681 | 0.17 |
ENST00000343003.10
ENST00000399949.6 |
SPATA13
|
spermatogenesis associated 13 |
| chr5_-_75717368 | 0.16 |
ENST00000514838.6
ENST00000506164.5 ENST00000502826.5 ENST00000428202.7 ENST00000503835.5 |
POC5
|
POC5 centriolar protein |
| chr18_-_55402187 | 0.16 |
ENST00000630268.2
ENST00000570177.6 |
TCF4
|
transcription factor 4 |
| chr10_+_24209129 | 0.16 |
ENST00000376454.8
ENST00000635504.1 |
KIAA1217
|
KIAA1217 |
| chr10_-_14548646 | 0.16 |
ENST00000378470.5
|
FAM107B
|
family with sequence similarity 107 member B |
| chr17_-_8118489 | 0.16 |
ENST00000380149.6
ENST00000448843.7 |
ALOXE3
|
arachidonate lipoxygenase 3 |
| chr6_-_89819699 | 0.16 |
ENST00000439638.1
ENST00000629399.2 ENST00000369393.8 |
MDN1
|
midasin AAA ATPase 1 |
| chr17_-_63435184 | 0.16 |
ENST00000585153.1
|
CYB561
|
cytochrome b561 |
| chr4_-_142305826 | 0.16 |
ENST00000514525.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase type II B |
| chr12_+_101594849 | 0.16 |
ENST00000547405.5
ENST00000452455.6 ENST00000392934.7 ENST00000547509.5 ENST00000361685.6 ENST00000549145.5 ENST00000361466.7 ENST00000553190.5 ENST00000545503.6 ENST00000536007.5 ENST00000541119.5 ENST00000551300.5 ENST00000550270.1 |
MYBPC1
|
myosin binding protein C1 |
| chr5_+_35856883 | 0.16 |
ENST00000506850.5
ENST00000303115.8 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr1_+_13416351 | 0.15 |
ENST00000602960.1
|
PRAMEF20
|
PRAME family member 20 |
| chr11_+_121590388 | 0.15 |
ENST00000527934.1
|
SORL1
|
sortilin related receptor 1 |
| chr8_-_143568854 | 0.15 |
ENST00000524906.5
ENST00000532862.1 ENST00000534459.5 |
MROH6
|
maestro heat like repeat family member 6 |
| chr19_+_57599874 | 0.15 |
ENST00000332854.11
ENST00000597700.6 ENST00000597864.1 |
ZNF530
|
zinc finger protein 530 |
| chr17_-_17582417 | 0.15 |
ENST00000395783.5
|
PEMT
|
phosphatidylethanolamine N-methyltransferase |
| chr14_-_50944428 | 0.15 |
ENST00000532462.5
|
PYGL
|
glycogen phosphorylase L |
| chr17_+_50165990 | 0.15 |
ENST00000262018.8
ENST00000344627.10 ENST00000682109.1 ENST00000504073.2 |
SGCA
|
sarcoglycan alpha |
| chr2_+_162344108 | 0.14 |
ENST00000437150.7
ENST00000453113.6 |
GCA
|
grancalcin |
| chr4_-_142305935 | 0.14 |
ENST00000511838.5
|
INPP4B
|
inositol polyphosphate-4-phosphatase type II B |
| chr5_+_140855882 | 0.14 |
ENST00000562220.2
ENST00000307360.6 ENST00000506939.6 |
PCDHA10
|
protocadherin alpha 10 |
| chr8_-_144416819 | 0.14 |
ENST00000526658.1
ENST00000301305.8 |
SLC39A4
|
solute carrier family 39 member 4 |
| chr8_-_144416481 | 0.14 |
ENST00000276833.9
|
SLC39A4
|
solute carrier family 39 member 4 |
| chr14_-_50944475 | 0.14 |
ENST00000216392.8
ENST00000544180.6 |
PYGL
|
glycogen phosphorylase L |
| chr10_+_88664439 | 0.13 |
ENST00000394375.7
ENST00000608620.5 ENST00000238983.9 ENST00000355843.2 |
LIPF
|
lipase F, gastric type |
| chr11_+_121576760 | 0.13 |
ENST00000532694.5
ENST00000534286.5 |
SORL1
|
sortilin related receptor 1 |
| chr2_+_162344338 | 0.13 |
ENST00000233612.8
|
GCA
|
grancalcin |
| chr21_+_42499600 | 0.13 |
ENST00000398341.7
|
SLC37A1
|
solute carrier family 37 member 1 |
| chr11_+_60280531 | 0.13 |
ENST00000532114.6
|
MS4A4A
|
membrane spanning 4-domains A4A |
| chr2_+_191678122 | 0.13 |
ENST00000425611.9
ENST00000410026.7 |
NABP1
|
nucleic acid binding protein 1 |
| chr3_-_151278535 | 0.13 |
ENST00000309170.8
|
P2RY14
|
purinergic receptor P2Y14 |
| chr14_+_61187544 | 0.13 |
ENST00000555185.5
ENST00000557294.5 ENST00000556778.5 |
PRKCH
|
protein kinase C eta |
| chr6_+_31670379 | 0.13 |
ENST00000409525.1
|
LY6G5B
|
lymphocyte antigen 6 family member G5B |
| chr14_+_94174284 | 0.13 |
ENST00000304338.8
|
PPP4R4
|
protein phosphatase 4 regulatory subunit 4 |
| chr1_-_25859352 | 0.13 |
ENST00000374298.4
ENST00000538789.5 |
AUNIP
|
aurora kinase A and ninein interacting protein |
| chr17_+_76384601 | 0.12 |
ENST00000592299.6
ENST00000590959.5 ENST00000323374.8 |
SPHK1
|
sphingosine kinase 1 |
| chr9_-_34376878 | 0.12 |
ENST00000297625.8
|
MYORG
|
myogenesis regulating glycosidase (putative) |
| chr19_-_48513161 | 0.12 |
ENST00000673139.1
|
LMTK3
|
lemur tyrosine kinase 3 |
| chr11_+_44726811 | 0.12 |
ENST00000533202.5
ENST00000520358.7 ENST00000533080.5 ENST00000520999.6 |
TSPAN18
|
tetraspanin 18 |
| chr8_-_144605699 | 0.12 |
ENST00000377307.6
ENST00000276826.5 |
ARHGAP39
|
Rho GTPase activating protein 39 |
| chr1_+_207321668 | 0.12 |
ENST00000367064.9
ENST00000314754.12 ENST00000367067.8 ENST00000644836.1 ENST00000343420.6 |
CD55
|
CD55 molecule (Cromer blood group) |
| chr4_+_15479204 | 0.12 |
ENST00000674945.1
ENST00000506643.5 ENST00000652443.1 ENST00000651385.1 |
CC2D2A
|
coiled-coil and C2 domain containing 2A |
| chr1_-_206921867 | 0.12 |
ENST00000628511.2
ENST00000367091.8 |
FCMR
|
Fc fragment of IgM receptor |
| chr6_-_47042260 | 0.12 |
ENST00000371243.2
|
ADGRF1
|
adhesion G protein-coupled receptor F1 |
| chr11_+_1834804 | 0.12 |
ENST00000341958.3
|
SYT8
|
synaptotagmin 8 |
| chr6_+_83853576 | 0.11 |
ENST00000369687.2
|
RIPPLY2
|
ripply transcriptional repressor 2 |
| chr4_+_8182066 | 0.11 |
ENST00000508641.2
|
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr1_+_86547070 | 0.11 |
ENST00000370563.3
|
CLCA4
|
chloride channel accessory 4 |
| chr19_-_15464525 | 0.11 |
ENST00000343625.12
|
RASAL3
|
RAS protein activator like 3 |
| chr6_-_143450662 | 0.11 |
ENST00000237283.9
|
ADAT2
|
adenosine deaminase tRNA specific 2 |
| chr6_+_31670167 | 0.11 |
ENST00000375864.4
|
LY6G5B
|
lymphocyte antigen 6 family member G5B |
| chr12_-_91058016 | 0.11 |
ENST00000266719.4
|
KERA
|
keratocan |
| chr2_+_90038848 | 0.11 |
ENST00000390270.2
|
IGKV3D-20
|
immunoglobulin kappa variable 3D-20 |
| chr13_+_42048645 | 0.11 |
ENST00000337343.9
ENST00000261491.9 ENST00000611224.1 |
DGKH
|
diacylglycerol kinase eta |
| chr1_+_207321532 | 0.10 |
ENST00000367063.6
ENST00000391921.9 ENST00000645323.1 |
CD55
|
CD55 molecule (Cromer blood group) |
| chr2_-_74529670 | 0.10 |
ENST00000377526.4
|
AUP1
|
AUP1 lipid droplet regulating VLDL assembly factor |
| chr22_+_22427517 | 0.10 |
ENST00000390300.2
|
IGLV5-37
|
immunoglobulin lambda variable 5-37 |
| chr12_-_112108743 | 0.10 |
ENST00000547133.1
ENST00000261745.9 |
NAA25
|
N-alpha-acetyltransferase 25, NatB auxiliary subunit |
| chr3_+_150546765 | 0.10 |
ENST00000406576.7
ENST00000460851.6 ENST00000482093.5 ENST00000273435.9 |
EIF2A
|
eukaryotic translation initiation factor 2A |
| chr16_-_84504612 | 0.10 |
ENST00000562447.5
ENST00000343629.11 ENST00000565765.1 |
MEAK7
|
MTOR associated protein, eak-7 homolog |
| chr7_-_5425404 | 0.10 |
ENST00000399434.2
|
TNRC18
|
trinucleotide repeat containing 18 |
| chr14_+_23372809 | 0.10 |
ENST00000397242.2
ENST00000329715.2 |
IL25
|
interleukin 25 |
| chr8_-_143939543 | 0.10 |
ENST00000345136.8
|
PLEC
|
plectin |
| chr11_+_34632464 | 0.10 |
ENST00000531794.5
|
EHF
|
ETS homologous factor |
| chr6_-_31120437 | 0.10 |
ENST00000376288.3
|
CDSN
|
corneodesmosin |
| chr19_-_1822038 | 0.10 |
ENST00000643515.1
|
REXO1
|
RNA exonuclease 1 homolog |
| chr15_-_28712323 | 0.10 |
ENST00000563027.1
|
GOLGA8M
|
golgin A8 family member M |
| chr16_+_30199860 | 0.10 |
ENST00000395138.6
|
SULT1A3
|
sulfotransferase family 1A member 3 |
| chrX_-_57680260 | 0.10 |
ENST00000434992.1
|
NLRP2B
|
NLR family pyrin domain containing 2B |
| chr3_+_150546671 | 0.10 |
ENST00000487799.5
|
EIF2A
|
eukaryotic translation initiation factor 2A |
| chr19_-_14674886 | 0.10 |
ENST00000344373.8
ENST00000595472.1 |
ADGRE3
|
adhesion G protein-coupled receptor E3 |
| chr2_-_88992903 | 0.10 |
ENST00000495489.1
|
IGKV1-8
|
immunoglobulin kappa variable 1-8 |
| chr22_-_26618026 | 0.10 |
ENST00000647684.1
|
CRYBB1
|
crystallin beta B1 |
| chr15_+_34225050 | 0.10 |
ENST00000559421.1
|
EMC4
|
ER membrane protein complex subunit 4 |
| chr9_+_69820799 | 0.09 |
ENST00000377197.8
|
C9orf135
|
chromosome 9 open reading frame 135 |
| chrX_-_20116871 | 0.09 |
ENST00000379651.7
ENST00000443379.7 ENST00000379643.10 |
MAP7D2
|
MAP7 domain containing 2 |
| chr7_+_77537258 | 0.09 |
ENST00000248594.11
|
PTPN12
|
protein tyrosine phosphatase non-receptor type 12 |
| chr1_-_204494752 | 0.09 |
ENST00000684373.1
|
PIK3C2B
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta |
| chr2_+_181986015 | 0.09 |
ENST00000409702.1
|
PPP1R1C
|
protein phosphatase 1 regulatory inhibitor subunit 1C |
| chr16_-_20327801 | 0.09 |
ENST00000381360.9
|
GP2
|
glycoprotein 2 |
| chr11_+_77128237 | 0.09 |
ENST00000409709.9
ENST00000620575.4 ENST00000458637.6 ENST00000409619.6 |
MYO7A
|
myosin VIIA |
| chr19_-_38930737 | 0.09 |
ENST00000430193.7
ENST00000600042.5 ENST00000221431.11 |
SARS2
|
seryl-tRNA synthetase 2, mitochondrial |
| chr17_-_2336435 | 0.09 |
ENST00000301364.10
ENST00000576112.2 |
TSR1
|
TSR1 ribosome maturation factor |
| chr12_+_57591158 | 0.09 |
ENST00000422156.7
ENST00000354947.10 ENST00000540759.6 ENST00000551772.5 ENST00000550465.5 |
PIP4K2C
|
phosphatidylinositol-5-phosphate 4-kinase type 2 gamma |
| chr11_-_47848467 | 0.09 |
ENST00000378460.6
|
NUP160
|
nucleoporin 160 |
| chr15_+_23565705 | 0.09 |
ENST00000568252.1
ENST00000649065.1 |
MKRN3
|
makorin ring finger protein 3 |
| chr11_-_85665077 | 0.09 |
ENST00000527447.2
|
CREBZF
|
CREB/ATF bZIP transcription factor |
| chr1_-_206921987 | 0.09 |
ENST00000530505.1
ENST00000442471.4 |
FCMR
|
Fc fragment of IgM receptor |
| chr11_+_75768718 | 0.09 |
ENST00000376262.7
|
DGAT2
|
diacylglycerol O-acyltransferase 2 |
| chr8_-_61646807 | 0.09 |
ENST00000522919.5
|
ASPH
|
aspartate beta-hydroxylase |
| chr13_-_96053370 | 0.09 |
ENST00000376712.4
ENST00000397618.7 ENST00000376747.8 ENST00000376714.7 ENST00000638479.1 ENST00000621375.5 |
UGGT2
|
UDP-glucose glycoprotein glucosyltransferase 2 |
| chr11_-_5227063 | 0.09 |
ENST00000335295.4
ENST00000485743.1 ENST00000647020.1 |
HBB
|
hemoglobin subunit beta |
| chr19_+_10625507 | 0.08 |
ENST00000590857.5
ENST00000588688.5 ENST00000586078.5 ENST00000335757.10 |
SLC44A2
|
solute carrier family 44 member 2 |
| chr21_+_31873010 | 0.08 |
ENST00000270112.7
|
HUNK
|
hormonally up-regulated Neu-associated kinase |
| chr11_-_126062782 | 0.08 |
ENST00000531738.6
|
CDON
|
cell adhesion associated, oncogene regulated |
| chr4_-_164383986 | 0.08 |
ENST00000507270.5
ENST00000514618.5 ENST00000503008.5 |
MARCHF1
|
membrane associated ring-CH-type finger 1 |
| chr1_-_114670018 | 0.08 |
ENST00000393274.6
ENST00000393276.7 |
DENND2C
|
DENN domain containing 2C |
| chr5_+_74685225 | 0.08 |
ENST00000261416.12
|
HEXB
|
hexosaminidase subunit beta |
| chr17_-_14236862 | 0.08 |
ENST00000420162.7
ENST00000431716.2 |
CDRT15
|
CMT1A duplicated region transcript 15 |
| chr18_-_55589795 | 0.08 |
ENST00000568740.5
ENST00000629387.2 |
TCF4
|
transcription factor 4 |
| chr18_-_55589836 | 0.08 |
ENST00000537578.5
ENST00000564403.6 |
TCF4
|
transcription factor 4 |
| chr10_+_102394488 | 0.08 |
ENST00000369966.8
|
NFKB2
|
nuclear factor kappa B subunit 2 |
| chr5_+_83077559 | 0.08 |
ENST00000511817.1
|
XRCC4
|
X-ray repair cross complementing 4 |
| chr11_+_27055215 | 0.08 |
ENST00000525090.1
|
BBOX1
|
gamma-butyrobetaine hydroxylase 1 |
| chr2_-_68952880 | 0.08 |
ENST00000481498.1
ENST00000328895.9 |
GKN2
|
gastrokine 2 |
| chr1_+_179082025 | 0.08 |
ENST00000367625.8
ENST00000367627.8 ENST00000352445.10 |
TOR3A
|
torsin family 3 member A |
| chr1_-_39639626 | 0.08 |
ENST00000372852.4
|
HEYL
|
hes related family bHLH transcription factor with YRPW motif like |
| chr18_+_63907948 | 0.08 |
ENST00000238508.8
|
SERPINB10
|
serpin family B member 10 |
| chr7_-_38249572 | 0.08 |
ENST00000436911.6
|
TRGC2
|
T cell receptor gamma constant 2 |
| chr11_-_47848313 | 0.08 |
ENST00000530326.5
ENST00000528071.5 |
NUP160
|
nucleoporin 160 |
| chr22_+_22409755 | 0.08 |
ENST00000390299.2
|
IGLV1-40
|
immunoglobulin lambda variable 1-40 |
| chr2_-_164841410 | 0.08 |
ENST00000342193.8
ENST00000375458.6 |
COBLL1
|
cordon-bleu WH2 repeat protein like 1 |
| chr1_+_44118813 | 0.08 |
ENST00000372299.4
|
KLF17
|
Kruppel like factor 17 |
| chr22_+_21632751 | 0.08 |
ENST00000292779.4
|
CCDC116
|
coiled-coil domain containing 116 |
| chr3_-_119660580 | 0.08 |
ENST00000493094.6
ENST00000264231.7 ENST00000468801.1 |
POPDC2
|
popeye domain containing 2 |
| chr15_-_58933668 | 0.08 |
ENST00000380516.7
|
SLTM
|
SAFB like transcription modulator |
| chr1_+_119368773 | 0.08 |
ENST00000457318.5
ENST00000622548.4 ENST00000325945.4 |
HAO2
|
hydroxyacid oxidase 2 |
| chr11_-_28108109 | 0.07 |
ENST00000263181.7
|
KIF18A
|
kinesin family member 18A |
| chr11_-_63015831 | 0.07 |
ENST00000430500.6
ENST00000336232.7 |
SLC22A8
|
solute carrier family 22 member 8 |
| chr17_-_3595831 | 0.07 |
ENST00000399759.7
|
TRPV1
|
transient receptor potential cation channel subfamily V member 1 |
| chr15_-_66565959 | 0.07 |
ENST00000537670.5
|
LCTL
|
lactase like |
| chr19_-_40390175 | 0.07 |
ENST00000291823.3
|
HIPK4
|
homeodomain interacting protein kinase 4 |
| chr1_-_155991198 | 0.07 |
ENST00000673475.1
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor 2 |
| chr8_-_30812867 | 0.07 |
ENST00000518243.5
|
PPP2CB
|
protein phosphatase 2 catalytic subunit beta |
| chr3_-_57544324 | 0.07 |
ENST00000495027.5
ENST00000351747.6 ENST00000389536.8 ENST00000311202.7 |
DNAH12
|
dynein axonemal heavy chain 12 |
| chr12_-_56636318 | 0.07 |
ENST00000549506.5
ENST00000379441.7 ENST00000551812.5 |
BAZ2A
|
bromodomain adjacent to zinc finger domain 2A |
| chr1_+_119368802 | 0.07 |
ENST00000361035.8
ENST00000419144.1 |
HAO2
HAO2-IT1
|
hydroxyacid oxidase 2 HAO2 intronic transcript 1 |
| chr2_-_232776555 | 0.07 |
ENST00000438786.1
ENST00000233826.4 ENST00000409779.1 |
KCNJ13
|
potassium inwardly rectifying channel subfamily J member 13 |
| chr2_+_85429448 | 0.07 |
ENST00000651736.1
|
SH2D6
|
SH2 domain containing 6 |
| chr3_+_50191604 | 0.07 |
ENST00000232461.8
ENST00000433068.5 |
GNAT1
|
G protein subunit alpha transducin 1 |
| chr2_+_231395702 | 0.07 |
ENST00000287590.6
|
B3GNT7
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7 |
| chr1_-_228406761 | 0.07 |
ENST00000366699.3
ENST00000284551.11 |
TRIM11
|
tripartite motif containing 11 |
| chr5_+_146447304 | 0.07 |
ENST00000296702.9
ENST00000394421.7 ENST00000679501.2 |
TCERG1
|
transcription elongation regulator 1 |
| chr14_+_39267377 | 0.07 |
ENST00000556148.5
ENST00000348007.7 |
MIA2
|
MIA SH3 domain ER export factor 2 |
| chr12_-_10098977 | 0.07 |
ENST00000315330.8
ENST00000457018.6 |
CLEC1A
|
C-type lectin domain family 1 member A |
| chr19_-_663174 | 0.07 |
ENST00000292363.10
ENST00000589762.5 |
RNF126
|
ring finger protein 126 |
| chrX_-_31072035 | 0.07 |
ENST00000359202.5
|
FTHL17
|
ferritin heavy chain like 17 |
| chr12_-_21941300 | 0.07 |
ENST00000684084.1
|
ABCC9
|
ATP binding cassette subfamily C member 9 |
| chr3_-_108589375 | 0.07 |
ENST00000625495.1
ENST00000619684.4 ENST00000295746.13 |
CIP2A
|
cellular inhibitor of PP2A |
| chr12_-_122730828 | 0.07 |
ENST00000432564.3
|
HCAR1
|
hydroxycarboxylic acid receptor 1 |
| chr2_-_85867641 | 0.07 |
ENST00000393808.8
|
ST3GAL5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MAFF_MAFG
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 0.3 | GO:0048560 | establishment of anatomical structure orientation(GO:0048560) |
| 0.1 | 0.3 | GO:1902769 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.1 | 0.3 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.1 | 0.7 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 0.4 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.1 | 0.3 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.1 | 0.3 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.1 | 0.2 | GO:1990654 | regulation of extrathymic T cell differentiation(GO:0033082) sebum secreting cell proliferation(GO:1990654) |
| 0.1 | 0.7 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.1 | 0.3 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 | 0.3 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.1 | 0.5 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.2 | GO:0009181 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.0 | 0.5 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.0 | 0.3 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.1 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.0 | 0.5 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.2 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.1 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.0 | 0.7 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.2 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.8 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.1 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.0 | 0.0 | GO:2000566 | positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.0 | 0.1 | GO:0050960 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.0 | 0.1 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.2 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.2 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.1 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.0 | 0.1 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.0 | 0.1 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0034350 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.0 | 0.3 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:0072014 | proximal tubule development(GO:0072014) |
| 0.0 | 0.1 | GO:0009624 | response to nematode(GO:0009624) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.0 | GO:0035552 | oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.0 | 0.8 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.0 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.0 | 0.0 | GO:1904021 | negative regulation of G-protein coupled receptor internalization(GO:1904021) |
| 0.0 | 0.0 | GO:0097045 | activation of blood coagulation via clotting cascade(GO:0002543) phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.0 | 0.2 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.0 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.0 | GO:0050883 | negative regulation of sodium:proton antiporter activity(GO:0032416) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 | 0.1 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.1 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.1 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.0 | 0.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.0 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.1 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 0.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.1 | 0.6 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.1 | 0.7 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.1 | 0.3 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.1 | 0.2 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.1 | 0.2 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.1 | 0.3 | GO:0008184 | purine nucleobase binding(GO:0002060) glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.5 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 0.3 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.1 | 0.2 | GO:0000773 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.0 | 0.3 | GO:0017161 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.0 | 0.1 | GO:0052852 | (S)-2-hydroxy-acid oxidase activity(GO:0003973) very-long-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052852) long-chain-(S)-2-hydroxy-long-chain-acid oxidase activity(GO:0052853) medium-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052854) |
| 0.0 | 0.5 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.2 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.0 | 1.0 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.0 | 0.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.2 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.4 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.0 | 0.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.6 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.8 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.5 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.1 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.0 | 0.1 | GO:0000035 | acyl binding(GO:0000035) |
| 0.0 | 0.1 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.2 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.0 | 0.1 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 0.0 | 0.1 | GO:0047291 | neolactotetraosylceramide alpha-2,3-sialyltransferase activity(GO:0004513) lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.0 | 0.0 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.2 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.2 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.0 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.0 | 0.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.1 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.9 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.5 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.6 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |