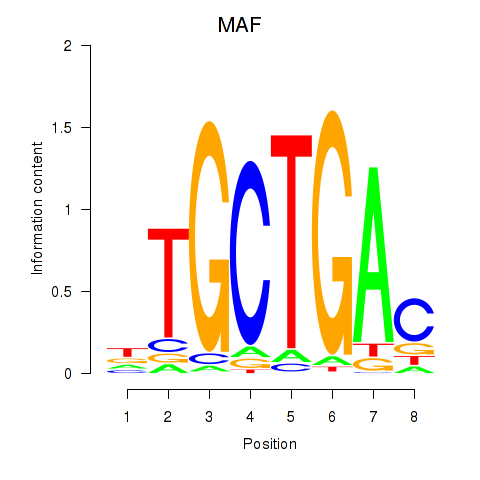
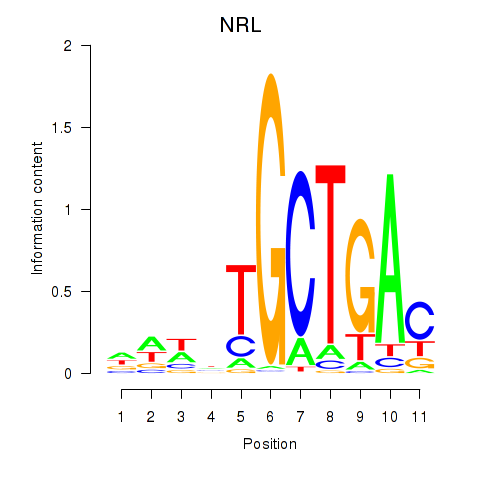
Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for MAF_NRL
Z-value: 0.32
Transcription factors associated with MAF_NRL
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MAF
|
ENSG00000178573.7 | MAF bZIP transcription factor |
|
NRL
|
ENSG00000129535.13 | neural retina leucine zipper |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NRL | hg38_v1_chr14_-_24081928_24081967, hg38_v1_chr14_-_24081986_24082005 | -0.36 | 3.9e-01 | Click! |
| MAF | hg38_v1_chr16_-_79599902_79600058 | -0.16 | 7.0e-01 | Click! |
Activity profile of MAF_NRL motif
Sorted Z-values of MAF_NRL motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_63887698 | 0.32 |
ENST00000457692.5
ENST00000299502.9 ENST00000413956.5 |
SERPINB2
|
serpin family B member 2 |
| chr1_-_201399525 | 0.32 |
ENST00000367313.4
|
LAD1
|
ladinin 1 |
| chr1_+_24319342 | 0.29 |
ENST00000361548.9
|
GRHL3
|
grainyhead like transcription factor 3 |
| chr1_+_24319511 | 0.29 |
ENST00000356046.6
|
GRHL3
|
grainyhead like transcription factor 3 |
| chr17_+_50532713 | 0.26 |
ENST00000503690.5
ENST00000514874.5 ENST00000268933.8 |
EPN3
|
epsin 3 |
| chr4_+_95051671 | 0.22 |
ENST00000440890.7
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr19_+_35115912 | 0.22 |
ENST00000603181.5
|
FXYD3
|
FXYD domain containing ion transport regulator 3 |
| chr7_-_41703062 | 0.19 |
ENST00000242208.5
|
INHBA
|
inhibin subunit beta A |
| chr1_-_201399302 | 0.19 |
ENST00000633953.1
ENST00000391967.7 |
LAD1
|
ladinin 1 |
| chr19_-_50953093 | 0.17 |
ENST00000593428.5
|
KLK5
|
kallikrein related peptidase 5 |
| chr19_-_50953063 | 0.16 |
ENST00000391809.6
|
KLK5
|
kallikrein related peptidase 5 |
| chr12_-_57767057 | 0.16 |
ENST00000228606.9
|
CYP27B1
|
cytochrome P450 family 27 subfamily B member 1 |
| chr2_+_206939515 | 0.16 |
ENST00000272852.4
|
CPO
|
carboxypeptidase O |
| chr9_-_33447553 | 0.16 |
ENST00000645858.1
ENST00000297991.6 |
AQP3
|
aquaporin 3 (Gill blood group) |
| chr1_+_2073462 | 0.16 |
ENST00000400921.6
|
PRKCZ
|
protein kinase C zeta |
| chrX_-_15664798 | 0.15 |
ENST00000380342.4
|
CLTRN
|
collectrin, amino acid transport regulator |
| chr12_-_52520371 | 0.15 |
ENST00000549420.1
ENST00000252242.9 ENST00000551275.1 ENST00000546577.1 |
KRT5
|
keratin 5 |
| chr7_+_26291850 | 0.14 |
ENST00000338523.9
ENST00000446848.6 |
SNX10
|
sorting nexin 10 |
| chr17_-_41612757 | 0.14 |
ENST00000301653.9
ENST00000593067.1 |
KRT16
|
keratin 16 |
| chr8_+_32721823 | 0.14 |
ENST00000539990.3
ENST00000519240.5 |
NRG1
|
neuregulin 1 |
| chr12_-_84911178 | 0.14 |
ENST00000681688.1
|
SLC6A15
|
solute carrier family 6 member 15 |
| chr11_+_70085413 | 0.14 |
ENST00000316296.9
ENST00000530676.5 |
ANO1
|
anoctamin 1 |
| chr1_+_2073986 | 0.13 |
ENST00000461106.6
|
PRKCZ
|
protein kinase C zeta |
| chr19_+_35115787 | 0.13 |
ENST00000604404.6
|
FXYD3
|
FXYD domain containing ion transport regulator 3 |
| chr19_+_35115872 | 0.13 |
ENST00000435734.6
|
FXYD3
|
FXYD domain containing ion transport regulator 3 |
| chrX_-_31178220 | 0.13 |
ENST00000681026.1
|
DMD
|
dystrophin |
| chrX_-_31178149 | 0.13 |
ENST00000679437.1
|
DMD
|
dystrophin |
| chr2_+_113127588 | 0.13 |
ENST00000409930.4
|
IL1RN
|
interleukin 1 receptor antagonist |
| chr17_-_78132407 | 0.13 |
ENST00000322914.7
|
TMC6
|
transmembrane channel like 6 |
| chr19_+_14941489 | 0.12 |
ENST00000248072.3
|
OR7C2
|
olfactory receptor family 7 subfamily C member 2 |
| chr17_-_58417547 | 0.12 |
ENST00000577716.5
|
RNF43
|
ring finger protein 43 |
| chr17_-_58417521 | 0.12 |
ENST00000584437.5
ENST00000407977.7 |
RNF43
|
ring finger protein 43 |
| chr11_-_118252279 | 0.12 |
ENST00000525386.5
ENST00000527472.1 ENST00000278949.9 |
MPZL3
|
myelin protein zero like 3 |
| chr14_+_23185316 | 0.12 |
ENST00000399910.5
ENST00000492621.5 |
RNF212B
|
ring finger protein 212B |
| chr2_-_142131004 | 0.12 |
ENST00000434794.1
ENST00000389484.8 |
LRP1B
|
LDL receptor related protein 1B |
| chr17_-_75878542 | 0.11 |
ENST00000254816.6
|
TRIM47
|
tripartite motif containing 47 |
| chr6_+_121435595 | 0.11 |
ENST00000649003.1
ENST00000282561.4 |
GJA1
|
gap junction protein alpha 1 |
| chr16_+_67199509 | 0.10 |
ENST00000477898.5
|
ELMO3
|
engulfment and cell motility 3 |
| chr11_+_6845683 | 0.10 |
ENST00000299454.5
|
OR10A5
|
olfactory receptor family 10 subfamily A member 5 |
| chr18_-_55586092 | 0.09 |
ENST00000563888.6
ENST00000540999.5 ENST00000627685.2 |
TCF4
|
transcription factor 4 |
| chr1_+_22007450 | 0.09 |
ENST00000400271.2
|
CELA3A
|
chymotrypsin like elastase 3A |
| chr3_+_189631373 | 0.09 |
ENST00000264731.8
ENST00000418709.6 ENST00000320472.9 ENST00000392460.7 ENST00000440651.6 |
TP63
|
tumor protein p63 |
| chr18_-_55422492 | 0.09 |
ENST00000561992.5
ENST00000630712.2 |
TCF4
|
transcription factor 4 |
| chr1_-_99766620 | 0.09 |
ENST00000646001.2
|
FRRS1
|
ferric chelate reductase 1 |
| chr14_+_23372809 | 0.09 |
ENST00000397242.2
ENST00000329715.2 |
IL25
|
interleukin 25 |
| chr18_-_55585773 | 0.09 |
ENST00000563824.5
ENST00000626425.2 ENST00000566514.5 ENST00000568673.5 ENST00000562847.5 ENST00000568147.5 |
TCF4
|
transcription factor 4 |
| chr3_+_50155024 | 0.09 |
ENST00000414301.5
ENST00000450338.5 ENST00000413852.5 |
SEMA3F
|
semaphorin 3F |
| chr10_+_69630227 | 0.09 |
ENST00000373279.6
|
FAM241B
|
family with sequence similarity 241 member B |
| chr18_-_55302613 | 0.08 |
ENST00000561831.7
|
TCF4
|
transcription factor 4 |
| chr1_-_12616762 | 0.08 |
ENST00000464917.5
|
DHRS3
|
dehydrogenase/reductase 3 |
| chr5_+_148826600 | 0.08 |
ENST00000305988.6
|
ADRB2
|
adrenoceptor beta 2 |
| chr14_-_23155302 | 0.08 |
ENST00000529705.6
|
SLC7A8
|
solute carrier family 7 member 8 |
| chr22_+_44677044 | 0.08 |
ENST00000006251.11
|
PRR5
|
proline rich 5 |
| chrX_+_106726663 | 0.08 |
ENST00000255499.3
|
RNF128
|
ring finger protein 128 |
| chr22_+_44677077 | 0.08 |
ENST00000403581.5
|
PRR5
|
proline rich 5 |
| chr8_+_103819244 | 0.08 |
ENST00000262231.14
ENST00000507740.5 ENST00000408894.6 |
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr22_+_44676808 | 0.08 |
ENST00000624862.3
|
PRR5
|
proline rich 5 |
| chr7_-_32071397 | 0.08 |
ENST00000396184.7
ENST00000396189.2 ENST00000321453.12 |
PDE1C
|
phosphodiesterase 1C |
| chr8_-_22133356 | 0.08 |
ENST00000680789.1
|
HR
|
HR lysine demethylase and nuclear receptor corepressor |
| chr12_+_20695323 | 0.08 |
ENST00000266509.7
|
SLCO1C1
|
solute carrier organic anion transporter family member 1C1 |
| chr19_-_55140922 | 0.07 |
ENST00000589745.5
|
TNNT1
|
troponin T1, slow skeletal type |
| chrX_+_37685773 | 0.07 |
ENST00000378616.5
|
XK
|
X-linked Kx blood group |
| chr7_-_22194709 | 0.07 |
ENST00000458533.5
|
RAPGEF5
|
Rap guanine nucleotide exchange factor 5 |
| chr5_+_67004618 | 0.07 |
ENST00000261569.11
ENST00000436277.5 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr14_+_21042352 | 0.07 |
ENST00000298690.5
|
RNASE7
|
ribonuclease A family member 7 |
| chr17_+_57105899 | 0.07 |
ENST00000576295.5
|
AKAP1
|
A-kinase anchoring protein 1 |
| chr19_-_55149193 | 0.07 |
ENST00000587758.5
ENST00000588981.6 ENST00000356783.9 ENST00000291901.12 ENST00000588426.5 ENST00000536926.5 ENST00000588147.5 |
TNNT1
|
troponin T1, slow skeletal type |
| chr8_-_74321532 | 0.07 |
ENST00000342232.5
|
JPH1
|
junctophilin 1 |
| chr18_+_11752041 | 0.07 |
ENST00000423027.8
|
GNAL
|
G protein subunit alpha L |
| chr19_-_51027662 | 0.07 |
ENST00000594768.5
|
KLK11
|
kallikrein related peptidase 11 |
| chr1_-_109393197 | 0.07 |
ENST00000538502.5
ENST00000482236.5 |
SORT1
|
sortilin 1 |
| chr1_+_61952036 | 0.06 |
ENST00000646453.1
ENST00000635137.1 |
PATJ
|
PATJ crumbs cell polarity complex component |
| chr3_+_50155305 | 0.06 |
ENST00000002829.8
ENST00000426511.5 |
SEMA3F
|
semaphorin 3F |
| chr6_-_11779606 | 0.06 |
ENST00000506810.1
|
ADTRP
|
androgen dependent TFPI regulating protein |
| chr9_+_98807619 | 0.06 |
ENST00000375011.4
|
GALNT12
|
polypeptide N-acetylgalactosaminyltransferase 12 |
| chr5_+_90474879 | 0.06 |
ENST00000504930.5
ENST00000514483.5 |
POLR3G
|
RNA polymerase III subunit G |
| chr1_-_115768702 | 0.06 |
ENST00000261448.6
|
CASQ2
|
calsequestrin 2 |
| chr15_-_59372863 | 0.06 |
ENST00000288235.9
|
MYO1E
|
myosin IE |
| chr1_+_9997220 | 0.06 |
ENST00000294435.8
|
RBP7
|
retinol binding protein 7 |
| chr18_-_55422306 | 0.06 |
ENST00000566777.5
ENST00000626584.2 |
TCF4
|
transcription factor 4 |
| chr2_+_188291661 | 0.06 |
ENST00000409843.5
|
GULP1
|
GULP PTB domain containing engulfment adaptor 1 |
| chr10_+_24466487 | 0.06 |
ENST00000396446.5
ENST00000396445.5 ENST00000376451.4 |
KIAA1217
|
KIAA1217 |
| chr2_+_188291854 | 0.06 |
ENST00000409830.6
|
GULP1
|
GULP PTB domain containing engulfment adaptor 1 |
| chrX_-_54798253 | 0.06 |
ENST00000218436.7
|
ITIH6
|
inter-alpha-trypsin inhibitor heavy chain family member 6 |
| chr9_-_114348966 | 0.06 |
ENST00000374079.8
|
AKNA
|
AT-hook transcription factor |
| chr5_+_55853314 | 0.06 |
ENST00000354961.8
ENST00000297015.7 |
IL31RA
|
interleukin 31 receptor A |
| chrY_-_2787676 | 0.06 |
ENST00000383070.2
|
SRY
|
sex determining region Y |
| chr2_+_188291994 | 0.05 |
ENST00000409927.5
ENST00000409805.5 |
GULP1
|
GULP PTB domain containing engulfment adaptor 1 |
| chr7_-_126533850 | 0.05 |
ENST00000444921.3
|
GRM8
|
glutamate metabotropic receptor 8 |
| chr10_-_49762276 | 0.05 |
ENST00000374103.9
|
OGDHL
|
oxoglutarate dehydrogenase L |
| chr9_-_109498251 | 0.05 |
ENST00000374541.4
ENST00000262539.7 |
PTPN3
|
protein tyrosine phosphatase non-receptor type 3 |
| chr14_-_23365149 | 0.05 |
ENST00000216733.8
|
EFS
|
embryonal Fyn-associated substrate |
| chr5_+_55853464 | 0.05 |
ENST00000652039.1
|
IL31RA
|
interleukin 31 receptor A |
| chrX_+_103215072 | 0.05 |
ENST00000372695.6
ENST00000372691.3 |
BEX4
|
brain expressed X-linked 4 |
| chr17_+_75754618 | 0.05 |
ENST00000584939.1
|
ITGB4
|
integrin subunit beta 4 |
| chr15_-_68206039 | 0.05 |
ENST00000395463.3
|
CALML4
|
calmodulin like 4 |
| chr17_+_57096572 | 0.05 |
ENST00000539273.5
|
AKAP1
|
A-kinase anchoring protein 1 |
| chr9_+_12693327 | 0.05 |
ENST00000388918.10
|
TYRP1
|
tyrosinase related protein 1 |
| chr12_+_7130341 | 0.05 |
ENST00000266546.11
|
CLSTN3
|
calsyntenin 3 |
| chr14_+_92652468 | 0.05 |
ENST00000556418.1
|
RIN3
|
Ras and Rab interactor 3 |
| chr10_-_49762335 | 0.05 |
ENST00000419399.4
ENST00000432695.2 |
OGDHL
|
oxoglutarate dehydrogenase L |
| chr2_-_64144411 | 0.05 |
ENST00000358912.5
|
PELI1
|
pellino E3 ubiquitin protein ligase 1 |
| chr22_+_20774092 | 0.05 |
ENST00000215727.10
|
SERPIND1
|
serpin family D member 1 |
| chrX_+_41447322 | 0.05 |
ENST00000378220.2
ENST00000342595.2 |
NYX
|
nyctalopin |
| chr7_-_19145306 | 0.05 |
ENST00000275461.3
|
FERD3L
|
Fer3 like bHLH transcription factor |
| chr6_-_52244500 | 0.05 |
ENST00000336123.5
|
IL17F
|
interleukin 17F |
| chr3_-_179243284 | 0.05 |
ENST00000486944.2
|
KCNMB3
|
potassium calcium-activated channel subfamily M regulatory beta subunit 3 |
| chr5_-_149063021 | 0.05 |
ENST00000515425.6
|
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr8_-_42377227 | 0.05 |
ENST00000220812.3
|
DKK4
|
dickkopf WNT signaling pathway inhibitor 4 |
| chr9_-_114682061 | 0.05 |
ENST00000612244.5
|
TEX48
|
testis expressed 48 |
| chr14_-_77320855 | 0.05 |
ENST00000556394.2
ENST00000261534.9 |
POMT2
|
protein O-mannosyltransferase 2 |
| chr10_-_96271508 | 0.05 |
ENST00000427367.6
ENST00000413476.6 ENST00000371176.6 |
BLNK
|
B cell linker |
| chr8_+_127735597 | 0.05 |
ENST00000651626.1
|
MYC
|
MYC proto-oncogene, bHLH transcription factor |
| chr6_-_42451910 | 0.05 |
ENST00000372922.8
ENST00000541110.5 |
TRERF1
|
transcriptional regulating factor 1 |
| chr9_+_135714582 | 0.05 |
ENST00000630792.2
|
KCNT1
|
potassium sodium-activated channel subfamily T member 1 |
| chr18_+_58195390 | 0.05 |
ENST00000456173.6
ENST00000676226.1 ENST00000675865.1 |
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr2_+_70900546 | 0.05 |
ENST00000234392.3
|
VAX2
|
ventral anterior homeobox 2 |
| chr15_+_80695277 | 0.05 |
ENST00000258884.5
ENST00000558464.1 |
ABHD17C
|
abhydrolase domain containing 17C, depalmitoylase |
| chr11_-_5243644 | 0.05 |
ENST00000643122.1
|
HBD
|
hemoglobin subunit delta |
| chr19_-_51024211 | 0.05 |
ENST00000593681.5
|
KLK11
|
kallikrein related peptidase 11 |
| chr17_-_40867200 | 0.05 |
ENST00000647902.1
ENST00000251643.5 |
KRT12
|
keratin 12 |
| chr19_-_50968125 | 0.05 |
ENST00000594641.1
|
KLK6
|
kallikrein related peptidase 6 |
| chr1_+_161721563 | 0.05 |
ENST00000367948.6
|
FCRLB
|
Fc receptor like B |
| chr17_-_29176752 | 0.05 |
ENST00000533112.5
|
MYO18A
|
myosin XVIIIA |
| chr2_-_74529670 | 0.04 |
ENST00000377526.4
|
AUP1
|
AUP1 lipid droplet regulating VLDL assembly factor |
| chr3_-_50303565 | 0.04 |
ENST00000266031.8
ENST00000395143.6 ENST00000457214.6 ENST00000447605.2 ENST00000395144.7 ENST00000418723.1 |
HYAL1
|
hyaluronidase 1 |
| chr14_-_77320741 | 0.04 |
ENST00000682795.1
ENST00000682247.1 |
POMT2
|
protein O-mannosyltransferase 2 |
| chr6_-_134052594 | 0.04 |
ENST00000275230.6
|
SLC2A12
|
solute carrier family 2 member 12 |
| chr6_-_33080710 | 0.04 |
ENST00000419277.5
|
HLA-DPA1
|
major histocompatibility complex, class II, DP alpha 1 |
| chr5_-_16936231 | 0.04 |
ENST00000507288.1
ENST00000274203.13 ENST00000513610.6 |
MYO10
|
myosin X |
| chr6_+_132538290 | 0.04 |
ENST00000434551.2
|
TAAR9
|
trace amine associated receptor 9 |
| chr2_+_236569817 | 0.04 |
ENST00000272928.4
|
ACKR3
|
atypical chemokine receptor 3 |
| chr17_+_29246852 | 0.04 |
ENST00000225387.8
|
CRYBA1
|
crystallin beta A1 |
| chr3_+_124584625 | 0.04 |
ENST00000291478.9
ENST00000682363.1 ENST00000454902.1 |
KALRN
|
kalirin RhoGEF kinase |
| chr22_+_41800662 | 0.04 |
ENST00000402061.7
ENST00000255784.6 |
CCDC134
|
coiled-coil domain containing 134 |
| chr7_+_142492121 | 0.04 |
ENST00000390374.3
|
TRBV7-6
|
T cell receptor beta variable 7-6 |
| chr4_+_77605807 | 0.04 |
ENST00000682537.1
|
CXCL13
|
C-X-C motif chemokine ligand 13 |
| chr9_+_135714425 | 0.04 |
ENST00000473941.5
ENST00000486577.6 |
KCNT1
|
potassium sodium-activated channel subfamily T member 1 |
| chr3_-_59049947 | 0.04 |
ENST00000491845.5
ENST00000472469.5 ENST00000482387.6 ENST00000295966.11 |
CFAP20DC
|
CFAP20 domain containing |
| chr10_-_96271553 | 0.04 |
ENST00000224337.10
|
BLNK
|
B cell linker |
| chr3_-_123620571 | 0.04 |
ENST00000583087.5
|
MYLK
|
myosin light chain kinase |
| chr12_-_70788914 | 0.04 |
ENST00000342084.8
|
PTPRR
|
protein tyrosine phosphatase receptor type R |
| chr6_+_31615215 | 0.04 |
ENST00000337917.11
ENST00000376059.8 |
AIF1
|
allograft inflammatory factor 1 |
| chr17_+_6641008 | 0.04 |
ENST00000570330.5
|
TXNDC17
|
thioredoxin domain containing 17 |
| chr9_+_27109393 | 0.04 |
ENST00000406359.8
|
TEK
|
TEK receptor tyrosine kinase |
| chr14_-_77320813 | 0.04 |
ENST00000682467.1
|
POMT2
|
protein O-mannosyltransferase 2 |
| chr21_+_43169008 | 0.04 |
ENST00000291554.6
|
CRYAA
|
crystallin alpha A |
| chr3_-_197226351 | 0.04 |
ENST00000656428.1
|
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr15_-_100341899 | 0.04 |
ENST00000568565.2
ENST00000268070.9 |
ADAMTS17
|
ADAM metallopeptidase with thrombospondin type 1 motif 17 |
| chr17_-_4545065 | 0.04 |
ENST00000572759.1
|
MYBBP1A
|
MYB binding protein 1a |
| chr8_+_131904071 | 0.04 |
ENST00000254624.10
ENST00000522709.5 |
EFR3A
|
EFR3 homolog A |
| chr12_-_91058016 | 0.04 |
ENST00000266719.4
|
KERA
|
keratocan |
| chr15_-_68205319 | 0.04 |
ENST00000467889.3
ENST00000448060.7 |
CALML4
|
calmodulin like 4 |
| chrX_+_129779930 | 0.04 |
ENST00000356892.4
|
SASH3
|
SAM and SH3 domain containing 3 |
| chr21_-_30216047 | 0.04 |
ENST00000399899.2
|
CLDN8
|
claudin 8 |
| chr12_+_20695553 | 0.04 |
ENST00000545102.1
|
SLCO1C1
|
solute carrier organic anion transporter family member 1C1 |
| chr16_-_81198766 | 0.04 |
ENST00000526632.5
|
PKD1L2
|
polycystin 1 like 2 (gene/pseudogene) |
| chr3_+_188947199 | 0.04 |
ENST00000433971.5
|
TPRG1
|
tumor protein p63 regulated 1 |
| chr19_+_40775511 | 0.04 |
ENST00000263369.4
ENST00000597140.5 |
MIA
|
MIA SH3 domain containing |
| chr14_-_105771405 | 0.04 |
ENST00000641136.1
ENST00000390551.6 |
IGHG3
|
immunoglobulin heavy constant gamma 3 (G3m marker) |
| chr2_+_134254065 | 0.04 |
ENST00000281923.4
|
MGAT5
|
alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase |
| chr8_+_122781621 | 0.03 |
ENST00000314393.6
|
ZHX2
|
zinc fingers and homeoboxes 2 |
| chr11_-_123885627 | 0.03 |
ENST00000528595.1
ENST00000375026.7 |
TMEM225
|
transmembrane protein 225 |
| chr2_+_195657179 | 0.03 |
ENST00000359634.10
|
SLC39A10
|
solute carrier family 39 member 10 |
| chr13_+_73058993 | 0.03 |
ENST00000377687.6
|
KLF5
|
Kruppel like factor 5 |
| chr11_+_118956289 | 0.03 |
ENST00000264031.3
|
UPK2
|
uroplakin 2 |
| chr3_-_190449782 | 0.03 |
ENST00000354905.3
|
TMEM207
|
transmembrane protein 207 |
| chr12_+_19205294 | 0.03 |
ENST00000424268.5
|
PLEKHA5
|
pleckstrin homology domain containing A5 |
| chr11_-_124441158 | 0.03 |
ENST00000328064.2
|
OR8B8
|
olfactory receptor family 8 subfamily B member 8 |
| chr19_+_40775154 | 0.03 |
ENST00000594436.5
ENST00000597784.5 |
MIA
|
MIA SH3 domain containing |
| chr11_+_63369779 | 0.03 |
ENST00000279178.4
|
SLC22A9
|
solute carrier family 22 member 9 |
| chr12_+_69825273 | 0.03 |
ENST00000547771.6
|
MYRFL
|
myelin regulatory factor like |
| chr19_-_3546306 | 0.03 |
ENST00000398558.8
|
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr11_+_77179750 | 0.03 |
ENST00000458169.2
|
MYO7A
|
myosin VIIA |
| chr6_+_33080445 | 0.03 |
ENST00000428835.5
|
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1 |
| chr4_-_6070162 | 0.03 |
ENST00000636216.1
ENST00000637373.2 |
ENSG00000284684.1
JAKMIP1
|
novel protein janus kinase and microtubule interacting protein 1 |
| chr21_+_29130630 | 0.03 |
ENST00000399926.5
ENST00000399928.6 |
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr1_+_32362537 | 0.03 |
ENST00000373534.4
|
TSSK3
|
testis specific serine kinase 3 |
| chr10_+_124461800 | 0.03 |
ENST00000368842.10
ENST00000392757.8 ENST00000368839.1 |
LHPP
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr9_+_2717479 | 0.03 |
ENST00000382082.4
|
KCNV2
|
potassium voltage-gated channel modifier subfamily V member 2 |
| chr2_+_74530018 | 0.03 |
ENST00000437202.1
|
HTRA2
|
HtrA serine peptidase 2 |
| chr22_-_26618026 | 0.03 |
ENST00000647684.1
|
CRYBB1
|
crystallin beta B1 |
| chr3_-_123620496 | 0.03 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr3_+_130850585 | 0.03 |
ENST00000505330.5
ENST00000504381.5 ENST00000507488.6 |
ATP2C1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr2_+_74529923 | 0.03 |
ENST00000258080.8
ENST00000352222.7 |
HTRA2
|
HtrA serine peptidase 2 |
| chr6_+_18387326 | 0.03 |
ENST00000259939.4
|
RNF144B
|
ring finger protein 144B |
| chr8_-_86743626 | 0.03 |
ENST00000320005.6
|
CNGB3
|
cyclic nucleotide gated channel subunit beta 3 |
| chr3_-_98522514 | 0.03 |
ENST00000503004.5
ENST00000506575.1 ENST00000513452.5 ENST00000515620.5 |
CLDND1
|
claudin domain containing 1 |
| chr19_+_48364361 | 0.03 |
ENST00000344846.7
|
SYNGR4
|
synaptogyrin 4 |
| chr11_+_62419025 | 0.03 |
ENST00000278282.3
|
SCGB1A1
|
secretoglobin family 1A member 1 |
| chr2_-_74441882 | 0.03 |
ENST00000272430.10
|
RTKN
|
rhotekin |
| chr16_+_46884323 | 0.03 |
ENST00000340124.9
|
GPT2
|
glutamic--pyruvic transaminase 2 |
| chr8_+_79611036 | 0.03 |
ENST00000220876.12
ENST00000518111.5 |
STMN2
|
stathmin 2 |
| chr10_+_119207560 | 0.03 |
ENST00000392870.3
|
GRK5
|
G protein-coupled receptor kinase 5 |
| chr8_-_20183127 | 0.03 |
ENST00000276373.10
ENST00000519026.5 ENST00000440926.3 ENST00000437980.3 |
SLC18A1
|
solute carrier family 18 member A1 |
| chr1_-_26354080 | 0.03 |
ENST00000308182.10
|
CRYBG2
|
crystallin beta-gamma domain containing 2 |
| chrX_-_155334580 | 0.03 |
ENST00000369449.7
ENST00000321926.4 |
CLIC2
|
chloride intracellular channel 2 |
| chr16_+_9091593 | 0.03 |
ENST00000327827.12
|
C16orf72
|
chromosome 16 open reading frame 72 |
| chr8_-_20183090 | 0.03 |
ENST00000265808.11
ENST00000522513.5 |
SLC18A1
|
solute carrier family 18 member A1 |
| chr10_+_113125536 | 0.03 |
ENST00000349937.7
|
TCF7L2
|
transcription factor 7 like 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MAF_NRL
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.2 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.3 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 0.2 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.3 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.0 | 0.2 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.2 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.0 | 0.6 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.1 | GO:0010652 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.0 | 0.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.1 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 | 0.2 | GO:0035290 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.0 | 0.3 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.1 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.0 | 0.0 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.1 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.0 | GO:0046247 | tetraterpenoid metabolic process(GO:0016108) carotenoid metabolic process(GO:0016116) carotene catabolic process(GO:0016121) xanthophyll metabolic process(GO:0016122) terpene catabolic process(GO:0046247) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.1 | GO:0009624 | response to nematode(GO:0009624) |
| 0.0 | 0.1 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.0 | GO:0090096 | regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.0 | 0.0 | GO:0035732 | nitric oxide storage(GO:0035732) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.1 | GO:0038129 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.1 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.0 | 0.0 | GO:1903937 | response to acrylamide(GO:1903937) |
| 0.0 | 0.0 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.0 | 0.1 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.3 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.3 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.0 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.1 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.0 | 0.0 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.0 | 0.1 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.2 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.0 | 0.4 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.1 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.0 | 0.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.0 | GO:0035730 | S-nitrosoglutathione binding(GO:0035730) dinitrosyl-iron complex binding(GO:0035731) |
| 0.0 | 0.0 | GO:0042282 | hydroxymethylglutaryl-CoA reductase (NADPH) activity(GO:0004420) hydroxymethylglutaryl-CoA reductase activity(GO:0042282) |
| 0.0 | 0.1 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.0 | 0.0 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0031014 | troponin T binding(GO:0031014) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID SHP2 PATHWAY | SHP2 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |