Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
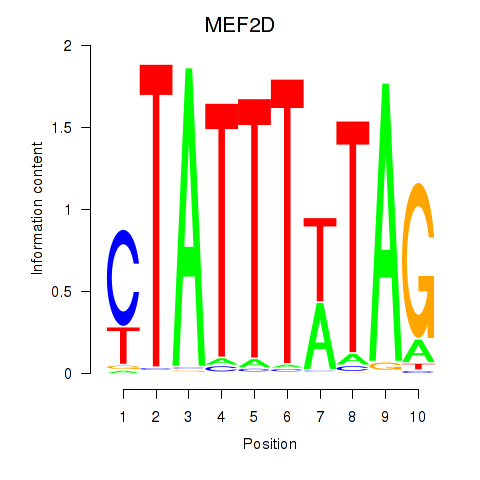
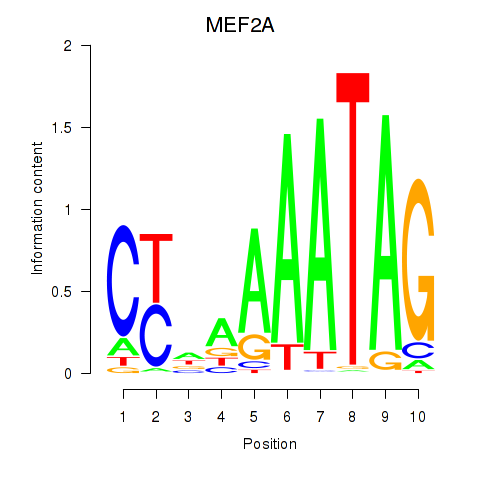
Results for MEF2D_MEF2A
Z-value: 1.05
Transcription factors associated with MEF2D_MEF2A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MEF2D
|
ENSG00000116604.18 | myocyte enhancer factor 2D |
|
MEF2A
|
ENSG00000068305.17 | myocyte enhancer factor 2A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MEF2D | hg38_v1_chr1_-_156500723_156500753 | -0.69 | 5.8e-02 | Click! |
| MEF2A | hg38_v1_chr15_+_99565921_99565929 | 0.54 | 1.7e-01 | Click! |
Activity profile of MEF2D_MEF2A motif
Sorted Z-values of MEF2D_MEF2A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_111756245 | 2.33 |
ENST00000447165.6
|
NREP
|
neuronal regeneration related protein |
| chr3_+_99638475 | 1.33 |
ENST00000452013.5
ENST00000261037.7 ENST00000652472.1 ENST00000273342.8 ENST00000621757.1 |
COL8A1
|
collagen type VIII alpha 1 chain |
| chr1_+_183636065 | 1.12 |
ENST00000304685.8
|
RGL1
|
ral guanine nucleotide dissociation stimulator like 1 |
| chr5_-_151141631 | 1.06 |
ENST00000523714.5
ENST00000521749.5 |
ANXA6
|
annexin A6 |
| chrX_-_143635081 | 0.99 |
ENST00000338017.8
|
SLITRK4
|
SLIT and NTRK like family member 4 |
| chr2_-_144431001 | 0.99 |
ENST00000636413.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr2_-_144430934 | 0.97 |
ENST00000638087.1
ENST00000638007.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr12_-_91146195 | 0.96 |
ENST00000548218.1
|
DCN
|
decorin |
| chr1_+_170664121 | 0.94 |
ENST00000239461.11
|
PRRX1
|
paired related homeobox 1 |
| chr2_-_189179754 | 0.86 |
ENST00000374866.9
ENST00000618828.1 |
COL5A2
|
collagen type V alpha 2 chain |
| chr1_+_170663917 | 0.73 |
ENST00000497230.2
|
PRRX1
|
paired related homeobox 1 |
| chr2_+_232633551 | 0.69 |
ENST00000264059.8
|
EFHD1
|
EF-hand domain family member D1 |
| chr6_-_154430495 | 0.67 |
ENST00000424998.3
|
CNKSR3
|
CNKSR family member 3 |
| chr1_+_113979391 | 0.62 |
ENST00000393300.6
ENST00000369551.5 |
OLFML3
|
olfactomedin like 3 |
| chr1_+_113979460 | 0.62 |
ENST00000320334.5
|
OLFML3
|
olfactomedin like 3 |
| chr6_+_135851681 | 0.55 |
ENST00000308191.11
|
PDE7B
|
phosphodiesterase 7B |
| chr2_+_148021404 | 0.54 |
ENST00000638043.2
|
MBD5
|
methyl-CpG binding domain protein 5 |
| chr3_-_112641128 | 0.52 |
ENST00000206423.8
|
CCDC80
|
coiled-coil domain containing 80 |
| chr6_+_131808011 | 0.50 |
ENST00000647893.1
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| chr1_-_145910031 | 0.47 |
ENST00000369304.8
|
ITGA10
|
integrin subunit alpha 10 |
| chr13_+_75804169 | 0.46 |
ENST00000526371.1
ENST00000526528.1 |
LMO7
|
LIM domain 7 |
| chr1_-_145910066 | 0.45 |
ENST00000539363.2
|
ITGA10
|
integrin subunit alpha 10 |
| chr3_-_112641292 | 0.45 |
ENST00000439685.6
|
CCDC80
|
coiled-coil domain containing 80 |
| chr9_+_121286115 | 0.45 |
ENST00000477104.2
ENST00000394353.7 |
GSN
|
gelsolin |
| chr5_+_93583212 | 0.44 |
ENST00000327111.8
|
NR2F1
|
nuclear receptor subfamily 2 group F member 1 |
| chr5_+_68288346 | 0.43 |
ENST00000320694.12
|
PIK3R1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr4_+_54229261 | 0.43 |
ENST00000508170.5
ENST00000512143.1 ENST00000257290.10 |
PDGFRA
|
platelet derived growth factor receptor alpha |
| chr5_+_173889337 | 0.42 |
ENST00000520867.5
ENST00000334035.9 |
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr4_-_39638893 | 0.42 |
ENST00000511809.5
ENST00000505729.1 |
SMIM14
|
small integral membrane protein 14 |
| chr3_+_138348570 | 0.41 |
ENST00000423968.7
|
MRAS
|
muscle RAS oncogene homolog |
| chr13_+_75804221 | 0.41 |
ENST00000489941.6
ENST00000525373.5 |
LMO7
|
LIM domain 7 |
| chr13_+_75760362 | 0.40 |
ENST00000534657.5
|
LMO7
|
LIM domain 7 |
| chr10_-_60944132 | 0.39 |
ENST00000337910.10
|
RHOBTB1
|
Rho related BTB domain containing 1 |
| chr3_+_138348592 | 0.37 |
ENST00000621127.4
ENST00000494949.5 |
MRAS
|
muscle RAS oncogene homolog |
| chr4_-_39638846 | 0.36 |
ENST00000295958.10
|
SMIM14
|
small integral membrane protein 14 |
| chr20_+_58907981 | 0.34 |
ENST00000656419.1
|
GNAS
|
GNAS complex locus |
| chrX_-_21758097 | 0.33 |
ENST00000379494.4
|
SMPX
|
small muscle protein X-linked |
| chr19_+_36605292 | 0.33 |
ENST00000460670.5
ENST00000292928.7 ENST00000439428.5 |
ZNF382
|
zinc finger protein 382 |
| chr3_+_69763726 | 0.33 |
ENST00000448226.9
|
MITF
|
melanocyte inducing transcription factor |
| chr16_+_88382950 | 0.32 |
ENST00000565624.3
|
ZNF469
|
zinc finger protein 469 |
| chr8_+_27325516 | 0.32 |
ENST00000346049.10
ENST00000420218.3 |
PTK2B
|
protein tyrosine kinase 2 beta |
| chr1_+_156126160 | 0.32 |
ENST00000448611.6
ENST00000368297.5 |
LMNA
|
lamin A/C |
| chr1_-_153545793 | 0.30 |
ENST00000354332.8
ENST00000368716.9 |
S100A4
|
S100 calcium binding protein A4 |
| chr8_-_27614681 | 0.29 |
ENST00000519472.5
ENST00000523589.5 ENST00000522413.5 ENST00000523396.1 ENST00000316403.15 |
CLU
|
clusterin |
| chr6_+_108656346 | 0.29 |
ENST00000540898.1
|
FOXO3
|
forkhead box O3 |
| chr21_-_26843063 | 0.29 |
ENST00000678221.1
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1 |
| chr1_-_31373067 | 0.28 |
ENST00000373713.7
|
FABP3
|
fatty acid binding protein 3 |
| chr21_-_26843012 | 0.28 |
ENST00000517777.6
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1 |
| chr17_+_70169516 | 0.28 |
ENST00000243457.4
|
KCNJ2
|
potassium inwardly rectifying channel subfamily J member 2 |
| chr17_+_44170695 | 0.28 |
ENST00000293414.6
|
ASB16
|
ankyrin repeat and SOCS box containing 16 |
| chr12_+_78864768 | 0.27 |
ENST00000261205.9
ENST00000457153.6 |
SYT1
|
synaptotagmin 1 |
| chr13_+_75760659 | 0.26 |
ENST00000526202.5
ENST00000465261.6 |
LMO7
|
LIM domain 7 |
| chr10_+_102419189 | 0.26 |
ENST00000432590.5
|
FBXL15
|
F-box and leucine rich repeat protein 15 |
| chr17_-_1485733 | 0.26 |
ENST00000648446.1
|
MYO1C
|
myosin IC |
| chr2_+_148021083 | 0.25 |
ENST00000642680.2
|
MBD5
|
methyl-CpG binding domain protein 5 |
| chr17_-_1486124 | 0.25 |
ENST00000575158.5
|
MYO1C
|
myosin IC |
| chr3_-_146161167 | 0.25 |
ENST00000360060.7
ENST00000282903.10 |
PLOD2
|
procollagen-lysine,2-oxoglutarate 5-dioxygenase 2 |
| chr8_-_13514821 | 0.25 |
ENST00000276297.9
|
DLC1
|
DLC1 Rho GTPase activating protein |
| chr7_-_134459089 | 0.25 |
ENST00000285930.9
|
AKR1B1
|
aldo-keto reductase family 1 member B |
| chr2_-_210303608 | 0.25 |
ENST00000341685.8
|
MYL1
|
myosin light chain 1 |
| chr9_+_136977496 | 0.25 |
ENST00000371625.8
ENST00000457950.5 |
PTGDS
|
prostaglandin D2 synthase |
| chr10_-_48652493 | 0.25 |
ENST00000435790.6
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr2_+_148021001 | 0.25 |
ENST00000407073.5
|
MBD5
|
methyl-CpG binding domain protein 5 |
| chr19_+_35139440 | 0.24 |
ENST00000455515.6
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr3_+_8501846 | 0.24 |
ENST00000454244.4
|
LMCD1
|
LIM and cysteine rich domains 1 |
| chr19_+_1266653 | 0.23 |
ENST00000586472.5
ENST00000589266.5 |
CIRBP
|
cold inducible RNA binding protein |
| chr4_+_128809684 | 0.23 |
ENST00000226319.11
ENST00000511647.5 |
JADE1
|
jade family PHD finger 1 |
| chr13_+_75636311 | 0.23 |
ENST00000377499.9
ENST00000377534.8 |
LMO7
|
LIM domain 7 |
| chr3_+_141387801 | 0.22 |
ENST00000514251.5
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr13_+_75760431 | 0.22 |
ENST00000321797.12
|
LMO7
|
LIM domain 7 |
| chr3_+_141387616 | 0.21 |
ENST00000509883.5
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr1_+_201648136 | 0.21 |
ENST00000367296.8
|
NAV1
|
neuron navigator 1 |
| chr2_+_219279330 | 0.21 |
ENST00000425450.5
ENST00000392086.8 ENST00000421532.5 ENST00000336576.10 |
DNAJB2
|
DnaJ heat shock protein family (Hsp40) member B2 |
| chr3_-_39192584 | 0.20 |
ENST00000340369.4
ENST00000421646.1 ENST00000396251.1 |
XIRP1
|
xin actin binding repeat containing 1 |
| chr10_-_3785179 | 0.20 |
ENST00000469435.1
|
KLF6
|
Kruppel like factor 6 |
| chr4_-_185775271 | 0.20 |
ENST00000430503.5
ENST00000319454.10 ENST00000450341.5 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr2_-_105438503 | 0.20 |
ENST00000393352.7
ENST00000607522.1 |
FHL2
|
four and a half LIM domains 2 |
| chr19_-_49423441 | 0.18 |
ENST00000270631.2
|
PTH2
|
parathyroid hormone 2 |
| chr8_-_13514744 | 0.18 |
ENST00000316609.9
|
DLC1
|
DLC1 Rho GTPase activating protein |
| chr3_+_8501807 | 0.18 |
ENST00000426878.2
ENST00000397386.7 ENST00000415597.5 ENST00000157600.8 |
LMCD1
|
LIM and cysteine rich domains 1 |
| chr22_+_19723525 | 0.18 |
ENST00000366425.4
|
GP1BB
|
glycoprotein Ib platelet subunit beta |
| chr1_+_156126525 | 0.17 |
ENST00000504687.6
ENST00000473598.6 |
LMNA
|
lamin A/C |
| chr3_-_138115594 | 0.17 |
ENST00000327532.7
ENST00000467030.5 |
DZIP1L
|
DAZ interacting zinc finger protein 1 like |
| chr17_-_36090133 | 0.16 |
ENST00000613922.2
|
CCL3
|
C-C motif chemokine ligand 3 |
| chr4_+_159241016 | 0.16 |
ENST00000644902.1
|
RAPGEF2
|
Rap guanine nucleotide exchange factor 2 |
| chr10_-_3785197 | 0.16 |
ENST00000497571.6
|
KLF6
|
Kruppel like factor 6 |
| chr3_+_138348823 | 0.16 |
ENST00000475711.5
ENST00000464896.5 |
MRAS
|
muscle RAS oncogene homolog |
| chr3_-_114624193 | 0.16 |
ENST00000481632.5
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr15_-_52652031 | 0.15 |
ENST00000546305.6
|
FAM214A
|
family with sequence similarity 214 member A |
| chr11_-_19201976 | 0.15 |
ENST00000647990.1
ENST00000649235.1 ENST00000265968.9 ENST00000649842.1 |
CSRP3
|
cysteine and glycine rich protein 3 |
| chr15_-_82806054 | 0.15 |
ENST00000541889.1
ENST00000334574.12 ENST00000561368.1 |
FSD2
|
fibronectin type III and SPRY domain containing 2 |
| chr5_-_67196791 | 0.15 |
ENST00000256447.5
|
CD180
|
CD180 molecule |
| chr1_-_46176482 | 0.15 |
ENST00000540385.2
ENST00000506599.2 |
P3R3URF-PIK3R3
P3R3URF
|
P3R3URF-PIK3R3 readthrough PIK3R3 upstream reading frame |
| chr2_-_210315160 | 0.15 |
ENST00000352451.4
|
MYL1
|
myosin light chain 1 |
| chr15_+_99565950 | 0.15 |
ENST00000557785.5
ENST00000558049.5 ENST00000449277.6 |
MEF2A
|
myocyte enhancer factor 2A |
| chr1_+_160081529 | 0.15 |
ENST00000368088.4
|
KCNJ9
|
potassium inwardly rectifying channel subfamily J member 9 |
| chr1_-_206772484 | 0.15 |
ENST00000423557.1
|
IL10
|
interleukin 10 |
| chr15_+_99565921 | 0.15 |
ENST00000558812.5
ENST00000338042.10 |
MEF2A
|
myocyte enhancer factor 2A |
| chr10_-_3785225 | 0.15 |
ENST00000542957.1
|
KLF6
|
Kruppel like factor 6 |
| chr2_+_44275457 | 0.15 |
ENST00000611973.4
ENST00000409387.5 |
SLC3A1
|
solute carrier family 3 member 1 |
| chr13_-_102759059 | 0.15 |
ENST00000322527.4
|
CCDC168
|
coiled-coil domain containing 168 |
| chr6_+_25726767 | 0.14 |
ENST00000274764.5
|
H2BC1
|
H2B clustered histone 1 |
| chr1_+_150272772 | 0.14 |
ENST00000369098.3
ENST00000369099.8 |
C1orf54
|
chromosome 1 open reading frame 54 |
| chr9_-_83267230 | 0.14 |
ENST00000328788.5
|
FRMD3
|
FERM domain containing 3 |
| chrX_-_47574738 | 0.14 |
ENST00000640721.1
|
SYN1
|
synapsin I |
| chr15_+_92900338 | 0.14 |
ENST00000625990.3
|
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr9_+_124291935 | 0.14 |
ENST00000546191.5
|
NEK6
|
NIMA related kinase 6 |
| chr8_-_69833338 | 0.14 |
ENST00000524945.5
|
SLCO5A1
|
solute carrier organic anion transporter family member 5A1 |
| chr1_-_58784035 | 0.14 |
ENST00000371222.4
|
JUN
|
Jun proto-oncogene, AP-1 transcription factor subunit |
| chr3_+_179653032 | 0.14 |
ENST00000680587.1
ENST00000681064.1 ENST00000263966.8 ENST00000681358.1 ENST00000679749.1 |
USP13
|
ubiquitin specific peptidase 13 |
| chr12_+_12785652 | 0.14 |
ENST00000356591.5
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr4_+_37891060 | 0.14 |
ENST00000261439.9
ENST00000508802.5 ENST00000402522.1 |
TBC1D1
|
TBC1 domain family member 1 |
| chr4_+_113049616 | 0.14 |
ENST00000504454.5
ENST00000357077.9 ENST00000394537.7 ENST00000672779.1 ENST00000264366.10 |
ANK2
|
ankyrin 2 |
| chr6_+_113857333 | 0.14 |
ENST00000612661.2
|
MARCKS
|
myristoylated alanine rich protein kinase C substrate |
| chr2_+_44275473 | 0.14 |
ENST00000260649.11
|
SLC3A1
|
solute carrier family 3 member 1 |
| chr15_+_99566066 | 0.14 |
ENST00000557942.5
|
MEF2A
|
myocyte enhancer factor 2A |
| chr8_-_27992624 | 0.13 |
ENST00000524352.5
|
SCARA5
|
scavenger receptor class A member 5 |
| chr4_-_113761068 | 0.13 |
ENST00000342666.9
ENST00000514328.5 ENST00000515496.5 ENST00000508738.5 ENST00000379773.6 |
CAMK2D
|
calcium/calmodulin dependent protein kinase II delta |
| chr3_-_177196451 | 0.13 |
ENST00000430069.5
ENST00000630796.2 ENST00000428970.5 |
TBL1XR1
|
TBL1X receptor 1 |
| chr11_-_64759967 | 0.13 |
ENST00000377432.7
ENST00000164139.4 |
PYGM
|
glycogen phosphorylase, muscle associated |
| chr1_+_89633086 | 0.13 |
ENST00000370454.9
|
LRRC8C
|
leucine rich repeat containing 8 VRAC subunit C |
| chr12_-_114683590 | 0.13 |
ENST00000257566.7
|
TBX3
|
T-box transcription factor 3 |
| chr7_-_151248668 | 0.13 |
ENST00000262188.13
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr12_+_15546344 | 0.12 |
ENST00000674388.1
ENST00000542557.5 ENST00000445537.6 ENST00000544244.5 ENST00000442921.7 |
PTPRO
|
protein tyrosine phosphatase receptor type O |
| chr2_-_165203870 | 0.12 |
ENST00000639244.1
ENST00000409101.7 ENST00000668657.1 |
SCN3A
|
sodium voltage-gated channel alpha subunit 3 |
| chr11_+_45896541 | 0.12 |
ENST00000395629.2
|
MAPK8IP1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr3_+_50279080 | 0.12 |
ENST00000316436.4
|
LSMEM2
|
leucine rich single-pass membrane protein 2 |
| chr3_+_179653129 | 0.12 |
ENST00000680008.1
|
USP13
|
ubiquitin specific peptidase 13 |
| chr9_+_35341830 | 0.12 |
ENST00000636694.1
|
UNC13B
|
unc-13 homolog B |
| chr14_-_88554898 | 0.12 |
ENST00000556564.6
|
PTPN21
|
protein tyrosine phosphatase non-receptor type 21 |
| chr15_+_90903288 | 0.12 |
ENST00000559717.6
|
MAN2A2
|
mannosidase alpha class 2A member 2 |
| chr16_-_15643024 | 0.12 |
ENST00000540441.6
|
MARF1
|
meiosis regulator and mRNA stability factor 1 |
| chr16_-_15643105 | 0.12 |
ENST00000548025.5
ENST00000551742.5 ENST00000396368.8 |
MARF1
|
meiosis regulator and mRNA stability factor 1 |
| chr4_-_185775890 | 0.12 |
ENST00000437304.6
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr3_+_54123452 | 0.11 |
ENST00000620722.4
ENST00000490478.5 |
CACNA2D3
|
calcium voltage-gated channel auxiliary subunit alpha2delta 3 |
| chr2_+_161136901 | 0.11 |
ENST00000259075.6
ENST00000432002.5 |
TANK
|
TRAF family member associated NFKB activator |
| chr2_+_42494547 | 0.11 |
ENST00000405592.5
|
MTA3
|
metastasis associated 1 family member 3 |
| chr17_+_7558296 | 0.11 |
ENST00000438470.5
ENST00000436057.5 |
TNFSF13
|
TNF superfamily member 13 |
| chr15_+_84817346 | 0.11 |
ENST00000258888.6
|
ALPK3
|
alpha kinase 3 |
| chr8_-_61689768 | 0.11 |
ENST00000517847.6
ENST00000389204.8 ENST00000517661.5 ENST00000517903.5 ENST00000522603.5 ENST00000541428.5 ENST00000522349.5 ENST00000522835.5 ENST00000518306.5 |
ASPH
|
aspartate beta-hydroxylase |
| chr17_+_82229034 | 0.11 |
ENST00000582743.6
ENST00000578684.6 ENST00000577650.6 ENST00000582715.1 |
SLC16A3
|
solute carrier family 16 member 3 |
| chr22_-_31346143 | 0.11 |
ENST00000405309.7
ENST00000351933.8 |
PATZ1
|
POZ/BTB and AT hook containing zinc finger 1 |
| chr1_+_84164962 | 0.11 |
ENST00000614872.4
ENST00000394839.6 |
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr2_-_165204042 | 0.10 |
ENST00000283254.12
ENST00000453007.1 |
SCN3A
|
sodium voltage-gated channel alpha subunit 3 |
| chr21_-_33643926 | 0.10 |
ENST00000438788.1
|
CRYZL1
|
crystallin zeta like 1 |
| chr22_+_25199856 | 0.10 |
ENST00000215855.7
ENST00000404334.1 |
CRYBB3
|
crystallin beta B3 |
| chr1_-_1000139 | 0.10 |
ENST00000428771.6
|
HES4
|
hes family bHLH transcription factor 4 |
| chr15_-_37099306 | 0.10 |
ENST00000557796.6
ENST00000397620.6 |
MEIS2
|
Meis homeobox 2 |
| chr1_+_151156659 | 0.10 |
ENST00000602841.5
|
SCNM1
|
sodium channel modifier 1 |
| chr2_+_161231078 | 0.10 |
ENST00000439442.1
|
TANK
|
TRAF family member associated NFKB activator |
| chr17_-_44123628 | 0.10 |
ENST00000587135.1
ENST00000225983.10 ENST00000682912.1 ENST00000588703.5 |
HDAC5
|
histone deacetylase 5 |
| chr6_+_122717544 | 0.09 |
ENST00000354275.2
ENST00000368446.1 |
PKIB
|
cAMP-dependent protein kinase inhibitor beta |
| chr19_+_35138993 | 0.09 |
ENST00000612146.4
ENST00000589209.5 |
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr17_+_39665340 | 0.09 |
ENST00000578283.1
ENST00000309889.3 |
TCAP
|
titin-cap |
| chr12_+_53461015 | 0.09 |
ENST00000553064.6
ENST00000547859.2 |
PCBP2
|
poly(rC) binding protein 2 |
| chr14_-_70080180 | 0.09 |
ENST00000394330.6
ENST00000533541.1 ENST00000216568.11 |
SLC8A3
|
solute carrier family 8 member A3 |
| chr3_-_158106408 | 0.09 |
ENST00000483851.7
|
SHOX2
|
short stature homeobox 2 |
| chr7_+_30911845 | 0.09 |
ENST00000652696.1
ENST00000311813.11 |
AQP1
|
aquaporin 1 (Colton blood group) |
| chrX_-_132219439 | 0.09 |
ENST00000370874.2
|
RAP2C
|
RAP2C, member of RAS oncogene family |
| chr8_-_71356653 | 0.09 |
ENST00000388742.8
ENST00000388740.4 |
EYA1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr10_-_102419934 | 0.09 |
ENST00000406432.5
|
PSD
|
pleckstrin and Sec7 domain containing |
| chrX_-_132219473 | 0.09 |
ENST00000620646.4
|
RAP2C
|
RAP2C, member of RAS oncogene family |
| chr4_+_119135825 | 0.09 |
ENST00000307128.6
|
MYOZ2
|
myozenin 2 |
| chr17_-_44123590 | 0.09 |
ENST00000336057.9
|
HDAC5
|
histone deacetylase 5 |
| chr2_+_167187283 | 0.09 |
ENST00000409605.1
ENST00000409273.6 |
XIRP2
|
xin actin binding repeat containing 2 |
| chr3_-_158106050 | 0.09 |
ENST00000441443.6
ENST00000389589.8 |
SHOX2
|
short stature homeobox 2 |
| chrX_-_15314543 | 0.09 |
ENST00000344384.8
|
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr6_-_2744126 | 0.08 |
ENST00000647417.1
|
MYLK4
|
myosin light chain kinase family member 4 |
| chr8_-_71356511 | 0.08 |
ENST00000419131.6
ENST00000388743.6 |
EYA1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr12_+_40224956 | 0.08 |
ENST00000298910.12
ENST00000343742.6 |
LRRK2
|
leucine rich repeat kinase 2 |
| chr12_+_68808143 | 0.08 |
ENST00000258149.11
ENST00000428863.6 ENST00000393412.7 |
MDM2
|
MDM2 proto-oncogene |
| chr9_+_126914760 | 0.08 |
ENST00000424082.6
ENST00000259351.10 ENST00000394022.7 ENST00000394011.7 ENST00000319107.8 |
RALGPS1
|
Ral GEF with PH domain and SH3 binding motif 1 |
| chr12_-_49187369 | 0.08 |
ENST00000547939.6
|
TUBA1A
|
tubulin alpha 1a |
| chr12_-_48999363 | 0.08 |
ENST00000421952.3
|
DDN
|
dendrin |
| chr19_+_10013468 | 0.08 |
ENST00000591589.3
|
RDH8
|
retinol dehydrogenase 8 |
| chr4_-_139177185 | 0.08 |
ENST00000394235.6
|
ELF2
|
E74 like ETS transcription factor 2 |
| chr9_-_33402551 | 0.08 |
ENST00000297988.6
ENST00000624075.3 ENST00000625032.1 ENST00000625109.3 |
AQP7
|
aquaporin 7 |
| chr2_-_230960954 | 0.08 |
ENST00000392039.2
|
GPR55
|
G protein-coupled receptor 55 |
| chr5_-_42811884 | 0.08 |
ENST00000514985.6
ENST00000511224.5 ENST00000507920.5 ENST00000510965.1 |
SELENOP
|
selenoprotein P |
| chr5_-_138139382 | 0.08 |
ENST00000265191.4
|
NME5
|
NME/NM23 family member 5 |
| chr9_-_127877665 | 0.08 |
ENST00000644144.2
|
AK1
|
adenylate kinase 1 |
| chr1_+_156242177 | 0.08 |
ENST00000368272.5
|
BGLAP
|
bone gamma-carboxyglutamate protein |
| chr16_+_8674605 | 0.08 |
ENST00000268251.13
|
ABAT
|
4-aminobutyrate aminotransferase |
| chr22_-_35617321 | 0.08 |
ENST00000397326.7
ENST00000442617.1 |
MB
|
myoglobin |
| chr5_+_161848112 | 0.08 |
ENST00000393943.10
|
GABRA1
|
gamma-aminobutyric acid type A receptor subunit alpha1 |
| chr5_-_141651376 | 0.08 |
ENST00000522783.5
ENST00000519800.1 ENST00000435817.7 |
FCHSD1
|
FCH and double SH3 domains 1 |
| chrX_-_72714181 | 0.08 |
ENST00000339490.7
ENST00000541944.5 ENST00000373539.3 |
PHKA1
|
phosphorylase kinase regulatory subunit alpha 1 |
| chr1_-_150720842 | 0.08 |
ENST00000442853.5
ENST00000368995.8 ENST00000322343.11 ENST00000361824.7 |
HORMAD1
|
HORMA domain containing 1 |
| chr6_-_43016856 | 0.08 |
ENST00000645375.1
ENST00000645410.1 |
MEA1
|
male-enhanced antigen 1 |
| chrX_-_72714278 | 0.08 |
ENST00000373542.9
ENST00000373545.7 |
PHKA1
|
phosphorylase kinase regulatory subunit alpha 1 |
| chr5_-_36151853 | 0.08 |
ENST00000296603.5
|
LMBRD2
|
LMBR1 domain containing 2 |
| chr2_+_167187364 | 0.08 |
ENST00000672671.1
|
XIRP2
|
xin actin binding repeat containing 2 |
| chr10_+_86668501 | 0.08 |
ENST00000623056.4
ENST00000263066.11 ENST00000429277.7 ENST00000361373.9 ENST00000372066.8 ENST00000372056.8 ENST00000623007.3 |
LDB3
|
LIM domain binding 3 |
| chr1_-_117210918 | 0.07 |
ENST00000369458.8
ENST00000430871.3 ENST00000328189.7 |
VTCN1
|
V-set domain containing T cell activation inhibitor 1 |
| chr11_-_18012883 | 0.07 |
ENST00000530613.5
ENST00000532389.5 ENST00000529728.5 ENST00000532265.5 |
SERGEF
|
secretion regulating guanine nucleotide exchange factor |
| chr1_-_203175783 | 0.07 |
ENST00000621380.1
ENST00000255416.9 |
MYBPH
|
myosin binding protein H |
| chr10_-_102419693 | 0.07 |
ENST00000611678.4
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr19_+_35138778 | 0.07 |
ENST00000351325.9
ENST00000586871.5 ENST00000592174.1 |
FXYD1
ENSG00000221857.7
|
FXYD domain containing ion transport regulator 1 novel transcript |
| chr4_-_82844418 | 0.07 |
ENST00000503937.5
|
SEC31A
|
SEC31 homolog A, COPII coat complex component |
| chrX_+_16650155 | 0.07 |
ENST00000380200.3
|
S100G
|
S100 calcium binding protein G |
Network of associatons between targets according to the STRING database.
First level regulatory network of MEF2D_MEF2A
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 2.0 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.2 | 1.0 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.2 | 0.5 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.1 | 0.4 | GO:0072275 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.1 | 0.3 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.1 | 1.4 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.1 | 0.3 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.1 | 0.3 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 0.3 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.1 | 0.4 | GO:1903906 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.1 | 0.3 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.1 | 0.5 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.1 | 0.2 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.1 | 0.4 | GO:0010734 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.1 | 0.3 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.1 | 0.2 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 0.1 | GO:0060932 | His-Purkinje system cell differentiation(GO:0060932) |
| 0.1 | 0.5 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.3 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.4 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.1 | 0.3 | GO:1902847 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.1 | 1.0 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.3 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 | 0.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 0.3 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.1 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.0 | 0.6 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.1 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.0 | 0.3 | GO:1990822 | L-cystine transport(GO:0015811) basic amino acid transmembrane transport(GO:1990822) |
| 0.0 | 0.2 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 1.2 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.2 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.0 | 0.2 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.1 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.1 | GO:0072019 | nitric oxide transport(GO:0030185) cerebrospinal fluid secretion(GO:0033326) proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) |
| 0.0 | 0.1 | GO:1903217 | negative regulation of late endosome to lysosome transport(GO:1902823) regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 0.0 | 0.1 | GO:1990785 | response to water-immersion restraint stress(GO:1990785) |
| 0.0 | 0.1 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.0 | 0.1 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.4 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.4 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.4 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.3 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.7 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.1 | GO:1904450 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.0 | 0.2 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:0032485 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.1 | GO:0046013 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.2 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.2 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.2 | GO:0036309 | protein localization to M-band(GO:0036309) |
| 0.0 | 0.1 | GO:0033594 | response to hydroxyisoflavone(GO:0033594) |
| 0.0 | 0.1 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.0 | 0.4 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.0 | GO:1905006 | negative regulation of alkaline phosphatase activity(GO:0010693) negative regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905006) |
| 0.0 | 0.1 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.1 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.1 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.1 | GO:0060631 | regulation of meiosis I(GO:0060631) |
| 0.0 | 0.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.2 | GO:2000172 | regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.0 | 0.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.0 | GO:2000547 | regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.0 | GO:0009305 | protein biotinylation(GO:0009305) histone biotinylation(GO:0071110) |
| 0.0 | 0.8 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.0 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.0 | 0.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0060613 | fat pad development(GO:0060613) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 0.9 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.1 | 1.3 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.1 | 0.4 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 0.5 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 1.0 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.3 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 0.3 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.1 | 0.3 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.5 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 1.0 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 1.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0044753 | amphisome(GO:0044753) |
| 0.0 | 0.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.1 | GO:0020003 | symbiont-containing vacuole(GO:0020003) |
| 0.0 | 0.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.4 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.9 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.5 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.3 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.1 | 0.3 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.1 | 0.2 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.1 | 0.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 1.8 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.3 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.3 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 1.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.3 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.5 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 1.0 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.3 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.2 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.0 | 0.2 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.2 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0032145 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.0 | 0.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 1.2 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.3 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.7 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.3 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 1.0 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.0 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.0 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.0 | GO:0004077 | biotin-[acetyl-CoA-carboxylase] ligase activity(GO:0004077) biotin-[methylcrotonoyl-CoA-carboxylase] ligase activity(GO:0004078) biotin-[methylmalonyl-CoA-carboxytransferase] ligase activity(GO:0004079) biotin-[propionyl-CoA-carboxylase (ATP-hydrolyzing)] ligase activity(GO:0004080) biotin-protein ligase activity(GO:0018271) |
| 0.0 | 0.1 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.9 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 1.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.8 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.5 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 1.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |