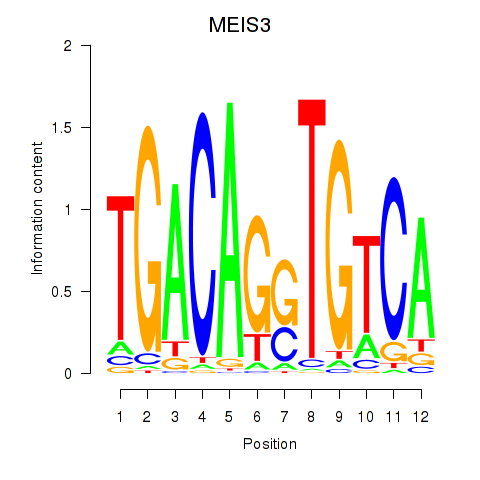
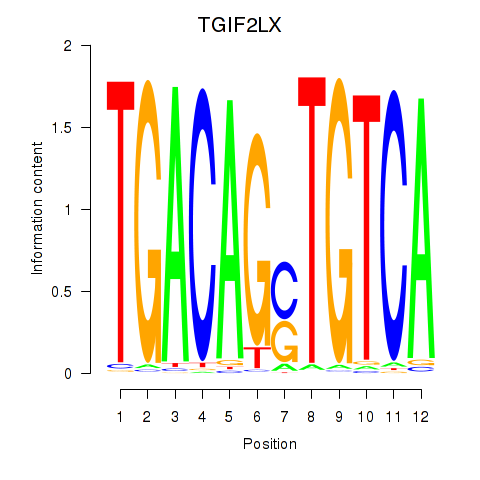
|
chr15_+_32717994
Show fit
|
0.56 |
ENST00000560677.5
ENST00000560830.1
ENST00000651154.1
|
GREM1
|
gremlin 1, DAN family BMP antagonist
|
|
chr15_+_32718476
Show fit
|
0.54 |
ENST00000652365.1
|
GREM1
|
gremlin 1, DAN family BMP antagonist
|
|
chr11_-_35526024
Show fit
|
0.48 |
ENST00000615849.4
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1
|
|
chr14_-_80211268
Show fit
|
0.38 |
ENST00000556811.5
|
DIO2
|
iodothyronine deiodinase 2
|
|
chr11_-_2149603
Show fit
|
0.30 |
ENST00000643349.1
|
ENSG00000284779.2
|
novel protein
|
|
chr6_-_52995170
Show fit
|
0.29 |
ENST00000370959.1
ENST00000370963.9
|
GSTA4
|
glutathione S-transferase alpha 4
|
|
chr1_+_201739864
Show fit
|
0.27 |
ENST00000367295.5
|
NAV1
|
neuron navigator 1
|
|
chr22_+_43923755
Show fit
|
0.25 |
ENST00000423180.2
ENST00000216180.8
|
PNPLA3
|
patatin like phospholipase domain containing 3
|
|
chr6_-_41705813
Show fit
|
0.24 |
ENST00000419574.6
ENST00000445214.2
|
TFEB
|
transcription factor EB
|
|
chr3_+_155083889
Show fit
|
0.24 |
ENST00000680282.1
|
MME
|
membrane metalloendopeptidase
|
|
chr11_+_120240135
Show fit
|
0.23 |
ENST00000543440.7
|
POU2F3
|
POU class 2 homeobox 3
|
|
chr17_-_19745602
Show fit
|
0.22 |
ENST00000444455.5
ENST00000439102.6
|
ALDH3A1
|
aldehyde dehydrogenase 3 family member A1
|
|
chr1_-_153544997
Show fit
|
0.22 |
ENST00000368715.5
|
S100A4
|
S100 calcium binding protein A4
|
|
chr3_+_155083523
Show fit
|
0.20 |
ENST00000680057.1
|
MME
|
membrane metalloendopeptidase
|
|
chr9_-_111038425
Show fit
|
0.20 |
ENST00000441240.1
ENST00000683809.1
|
LPAR1
|
lysophosphatidic acid receptor 1
|
|
chr1_+_159780930
Show fit
|
0.20 |
ENST00000368109.5
ENST00000368108.7
ENST00000368107.2
|
DUSP23
|
dual specificity phosphatase 23
|
|
chr12_+_104064520
Show fit
|
0.19 |
ENST00000229330.9
|
HCFC2
|
host cell factor C2
|
|
chr3_-_49689053
Show fit
|
0.19 |
ENST00000449682.2
|
MST1
|
macrophage stimulating 1
|
|
chr1_+_78649818
Show fit
|
0.16 |
ENST00000370747.9
ENST00000438486.1
|
IFI44
|
interferon induced protein 44
|
|
chr7_+_134646798
Show fit
|
0.14 |
ENST00000418040.5
ENST00000393132.2
|
BPGM
|
bisphosphoglycerate mutase
|
|
chr2_+_10420021
Show fit
|
0.14 |
ENST00000422133.1
|
HPCAL1
|
hippocalcin like 1
|
|
chr16_+_88382950
Show fit
|
0.14 |
ENST00000565624.3
|
ZNF469
|
zinc finger protein 469
|
|
chr2_+_24491860
Show fit
|
0.14 |
ENST00000406961.5
ENST00000405141.5
|
NCOA1
|
nuclear receptor coactivator 1
|
|
chr7_+_134646845
Show fit
|
0.14 |
ENST00000344924.8
|
BPGM
|
bisphosphoglycerate mutase
|
|
chr10_-_103452384
Show fit
|
0.14 |
ENST00000369788.7
|
CALHM2
|
calcium homeostasis modulator family member 2
|
|
chr3_-_3179674
Show fit
|
0.14 |
ENST00000424814.5
ENST00000450014.1
ENST00000231948.9
ENST00000432408.6
ENST00000639284.1
|
CRBN
|
cereblon
|
|
chr17_-_35119801
Show fit
|
0.13 |
ENST00000592577.5
ENST00000590016.5
ENST00000345365.11
|
RAD51D
|
RAD51 paralog D
|
|
chr14_-_80211472
Show fit
|
0.13 |
ENST00000557125.1
ENST00000438257.9
ENST00000422005.7
|
DIO2
|
iodothyronine deiodinase 2
|
|
chr1_-_47190013
Show fit
|
0.13 |
ENST00000294338.7
|
PDZK1IP1
|
PDZK1 interacting protein 1
|
|
chr17_+_60677822
Show fit
|
0.12 |
ENST00000407086.8
ENST00000589222.5
ENST00000626960.2
ENST00000390652.9
|
BCAS3
|
BCAS3 microtubule associated cell migration factor
|
|
chr4_+_677922
Show fit
|
0.12 |
ENST00000400159.6
|
MYL5
|
myosin light chain 5
|
|
chr6_+_24494939
Show fit
|
0.12 |
ENST00000348925.2
ENST00000357578.8
|
ALDH5A1
|
aldehyde dehydrogenase 5 family member A1
|
|
chr9_-_96778053
Show fit
|
0.12 |
ENST00000375231.5
ENST00000223428.9
|
ZNF510
|
zinc finger protein 510
|
|
chr10_-_103452356
Show fit
|
0.12 |
ENST00000260743.10
|
CALHM2
|
calcium homeostasis modulator family member 2
|
|
chr19_-_8981342
Show fit
|
0.12 |
ENST00000397910.8
|
MUC16
|
mucin 16, cell surface associated
|
|
chr17_-_35119733
Show fit
|
0.12 |
ENST00000460118.6
ENST00000335858.11
|
RAD51D
|
RAD51 paralog D
|
|
chr18_+_3449413
Show fit
|
0.11 |
ENST00000549253.5
|
TGIF1
|
TGFB induced factor homeobox 1
|
|
chr19_+_12163049
Show fit
|
0.11 |
ENST00000425827.5
ENST00000439995.5
ENST00000652580.1
ENST00000343979.6
ENST00000418338.1
|
ZNF136
|
zinc finger protein 136
|
|
chr11_+_68008542
Show fit
|
0.11 |
ENST00000614849.4
|
ALDH3B1
|
aldehyde dehydrogenase 3 family member B1
|
|
chr16_+_20806517
Show fit
|
0.10 |
ENST00000348433.10
ENST00000568501.5
ENST00000261377.11
ENST00000566276.5
|
REXO5
|
RNA exonuclease 5
|
|
chr2_-_65366650
Show fit
|
0.10 |
ENST00000443619.6
|
SPRED2
|
sprouty related EVH1 domain containing 2
|
|
chr7_+_76510516
Show fit
|
0.10 |
ENST00000257632.9
|
UPK3B
|
uroplakin 3B
|
|
chr17_-_35089212
Show fit
|
0.10 |
ENST00000584655.5
ENST00000447669.6
ENST00000315249.11
|
RFFL
|
ring finger and FYVE like domain containing E3 ubiquitin protein ligase
|
|
chr22_-_31489528
Show fit
|
0.10 |
ENST00000397525.5
|
EIF4ENIF1
|
eukaryotic translation initiation factor 4E nuclear import factor 1
|
|
chr9_-_22009272
Show fit
|
0.10 |
ENST00000380142.5
ENST00000276925.7
|
CDKN2B
|
cyclin dependent kinase inhibitor 2B
|
|
chr16_-_2210833
Show fit
|
0.10 |
ENST00000562360.5
ENST00000328540.8
ENST00000566018.1
|
BRICD5
|
BRICHOS domain containing 5
|
|
chr3_+_29280945
Show fit
|
0.10 |
ENST00000434693.6
|
RBMS3
|
RNA binding motif single stranded interacting protein 3
|
|
chr3_-_187745460
Show fit
|
0.09 |
ENST00000406870.7
|
BCL6
|
BCL6 transcription repressor
|
|
chr7_+_76510528
Show fit
|
0.09 |
ENST00000334348.8
|
UPK3B
|
uroplakin 3B
|
|
chr10_-_7619660
Show fit
|
0.09 |
ENST00000613909.4
|
ITIH5
|
inter-alpha-trypsin inhibitor heavy chain 5
|
|
chr11_+_123590939
Show fit
|
0.09 |
ENST00000646146.1
|
GRAMD1B
|
GRAM domain containing 1B
|
|
chr5_+_102808057
Show fit
|
0.09 |
ENST00000684043.1
ENST00000682407.1
|
PAM
|
peptidylglycine alpha-amidating monooxygenase
|
|
chrX_+_154547606
Show fit
|
0.09 |
ENST00000594239.6
ENST00000615874.4
ENST00000619941.4
ENST00000617207.4
ENST00000611176.4
|
IKBKG
|
inhibitor of nuclear factor kappa B kinase regulatory subunit gamma
|
|
chr10_+_102743938
Show fit
|
0.09 |
ENST00000448841.7
|
WBP1L
|
WW domain binding protein 1 like
|
|
chr4_+_139454070
Show fit
|
0.09 |
ENST00000305626.6
|
RAB33B
|
RAB33B, member RAS oncogene family
|
|
chr10_+_102776237
Show fit
|
0.09 |
ENST00000369889.5
|
WBP1L
|
WW domain binding protein 1 like
|
|
chr3_-_71306012
Show fit
|
0.08 |
ENST00000649431.1
ENST00000610810.5
|
FOXP1
|
forkhead box P1
|
|
chr10_-_27243019
Show fit
|
0.08 |
ENST00000679293.1
|
ACBD5
|
acyl-CoA binding domain containing 5
|
|
chr7_-_128775793
Show fit
|
0.08 |
ENST00000249389.3
|
OPN1SW
|
opsin 1, short wave sensitive
|
|
chr5_+_15500172
Show fit
|
0.08 |
ENST00000504595.2
|
FBXL7
|
F-box and leucine rich repeat protein 7
|
|
chr4_-_18021727
Show fit
|
0.08 |
ENST00000675605.1
ENST00000675927.1
ENST00000674942.1
ENST00000675143.1
ENST00000382226.5
ENST00000326877.8
ENST00000635767.1
|
LCORL
|
ligand dependent nuclear receptor corepressor like
|
|
chr16_+_20806698
Show fit
|
0.08 |
ENST00000564274.5
ENST00000563068.1
|
REXO5
|
RNA exonuclease 5
|
|
chr1_-_145885826
Show fit
|
0.08 |
ENST00000544626.2
ENST00000355594.9
|
ANKRD35
|
ankyrin repeat domain 35
|
|
chr4_-_119300546
Show fit
|
0.08 |
ENST00000504110.2
|
C4orf3
|
chromosome 4 open reading frame 3
|
|
chr16_+_56336767
Show fit
|
0.08 |
ENST00000640469.1
|
GNAO1
|
G protein subunit alpha o1
|
|
chr3_-_71305986
Show fit
|
0.07 |
ENST00000647614.1
|
FOXP1
|
forkhead box P1
|
|
chr7_+_66740725
Show fit
|
0.07 |
ENST00000442563.5
ENST00000450873.6
ENST00000284957.9
|
RABGEF1
|
RAB guanine nucleotide exchange factor 1
|
|
chr17_+_50095285
Show fit
|
0.07 |
ENST00000503614.5
|
PDK2
|
pyruvate dehydrogenase kinase 2
|
|
chrX_+_149540968
Show fit
|
0.07 |
ENST00000423421.5
ENST00000423540.6
ENST00000434353.6
ENST00000393985.8
ENST00000514208.5
|
EOLA1
|
endothelium and lymphocyte associated ASCH domain 1
|
|
chr18_+_79069385
Show fit
|
0.07 |
ENST00000426216.6
ENST00000307671.12
ENST00000586672.5
ENST00000586722.5
|
ATP9B
|
ATPase phospholipid transporting 9B (putative)
|
|
chr12_-_56221701
Show fit
|
0.07 |
ENST00000615206.4
ENST00000549038.5
ENST00000552244.5
|
RNF41
|
ring finger protein 41
|
|
chr15_-_26939518
Show fit
|
0.07 |
ENST00000541819.6
|
GABRB3
|
gamma-aminobutyric acid type A receptor subunit beta3
|
|
chr3_-_52685794
Show fit
|
0.07 |
ENST00000424867.1
ENST00000394830.7
ENST00000431678.5
ENST00000450271.5
|
PBRM1
|
polybromo 1
|
|
chr19_+_5455409
Show fit
|
0.07 |
ENST00000222033.6
|
ZNRF4
|
zinc and ring finger 4
|
|
chrX_-_13817346
Show fit
|
0.07 |
ENST00000356942.9
|
GPM6B
|
glycoprotein M6B
|
|
chr2_+_161231078
Show fit
|
0.07 |
ENST00000439442.1
|
TANK
|
TRAF family member associated NFKB activator
|
|
chr11_+_827545
Show fit
|
0.07 |
ENST00000528542.6
|
CRACR2B
|
calcium release activated channel regulator 2B
|
|
chr3_-_64268161
Show fit
|
0.07 |
ENST00000564377.6
|
PRICKLE2
|
prickle planar cell polarity protein 2
|
|
chr11_-_842508
Show fit
|
0.07 |
ENST00000322028.5
|
POLR2L
|
RNA polymerase II, I and III subunit L
|
|
chr19_-_49896868
Show fit
|
0.07 |
ENST00000593956.5
ENST00000391826.7
|
IL4I1
|
interleukin 4 induced 1
|
|
chr16_+_56336805
Show fit
|
0.06 |
ENST00000564727.2
|
GNAO1
|
G protein subunit alpha o1
|
|
chr20_-_58515382
Show fit
|
0.06 |
ENST00000371149.8
|
APCDD1L
|
APC down-regulated 1 like
|
|
chr16_+_1240698
Show fit
|
0.06 |
ENST00000561736.2
ENST00000338844.8
ENST00000461509.6
|
TPSAB1
|
tryptase alpha/beta 1
|
|
chr12_-_56221909
Show fit
|
0.06 |
ENST00000394013.6
ENST00000345093.9
ENST00000551711.5
ENST00000552656.5
|
RNF41
|
ring finger protein 41
|
|
chr2_+_148644706
Show fit
|
0.06 |
ENST00000258484.11
|
EPC2
|
enhancer of polycomb homolog 2
|
|
chr1_-_155192867
Show fit
|
0.06 |
ENST00000342482.8
ENST00000343256.9
ENST00000368389.6
ENST00000368390.7
ENST00000368396.8
ENST00000368398.7
ENST00000471283.5
ENST00000337604.6
ENST00000368392.7
ENST00000368393.7
ENST00000438413.5
ENST00000457295.6
ENST00000462215.5
ENST00000620103.4
ENST00000338684.9
ENST00000610359.4
ENST00000611571.4
ENST00000611577.4
ENST00000612778.4
ENST00000614519.4
ENST00000615517.4
|
MUC1
|
mucin 1, cell surface associated
|
|
chrX_+_149540593
Show fit
|
0.06 |
ENST00000450602.6
ENST00000441248.5
|
EOLA1
|
endothelium and lymphocyte associated ASCH domain 1
|
|
chr7_+_76510608
Show fit
|
0.06 |
ENST00000394849.1
|
UPK3B
|
uroplakin 3B
|
|
chr19_-_12551426
Show fit
|
0.06 |
ENST00000416136.1
ENST00000339282.12
ENST00000596193.1
ENST00000428311.1
|
ZNF564
ENSG00000196826.7
|
zinc finger protein 564
novel zinc finger protein
|
|
chr10_-_31031911
Show fit
|
0.06 |
ENST00000375311.1
ENST00000413025.5
ENST00000436087.6
ENST00000452305.5
ENST00000442986.5
|
ZNF438
|
zinc finger protein 438
|
|
chr3_+_131026844
Show fit
|
0.06 |
ENST00000510769.5
ENST00000383366.9
ENST00000510688.5
ENST00000511262.5
|
NEK11
|
NIMA related kinase 11
|
|
chr2_+_241150418
Show fit
|
0.06 |
ENST00000404405.7
ENST00000439916.5
ENST00000234038.11
ENST00000406106.7
ENST00000401987.5
|
PPP1R7
|
protein phosphatase 1 regulatory subunit 7
|
|
chr11_-_78417788
Show fit
|
0.06 |
ENST00000361507.5
|
GAB2
|
GRB2 associated binding protein 2
|
|
chr9_-_112890868
Show fit
|
0.06 |
ENST00000374228.5
|
SLC46A2
|
solute carrier family 46 member 2
|
|
chr12_-_10849464
Show fit
|
0.06 |
ENST00000544994.5
ENST00000228811.8
ENST00000540107.2
|
PRR4
|
proline rich 4
|
|
chr1_+_116111395
Show fit
|
0.06 |
ENST00000684484.1
ENST00000369500.4
|
MAB21L3
|
mab-21 like 3
|
|
chr11_+_828150
Show fit
|
0.06 |
ENST00000450448.5
|
CRACR2B
|
calcium release activated channel regulator 2B
|
|
chr2_+_241149631
Show fit
|
0.05 |
ENST00000407025.5
ENST00000272983.12
|
PPP1R7
|
protein phosphatase 1 regulatory subunit 7
|
|
chr9_-_83817632
Show fit
|
0.05 |
ENST00000376365.7
ENST00000376371.7
|
GKAP1
|
G kinase anchoring protein 1
|
|
chr16_-_18375069
Show fit
|
0.05 |
ENST00000545114.5
|
NPIPA9
|
nuclear pore complex interacting protein family, member A9
|
|
chr4_-_21697755
Show fit
|
0.05 |
ENST00000382148.7
|
KCNIP4
|
potassium voltage-gated channel interacting protein 4
|
|
chr10_-_102837406
Show fit
|
0.05 |
ENST00000369887.4
ENST00000638272.1
ENST00000639393.1
ENST00000638971.1
ENST00000638190.1
|
CYP17A1
|
cytochrome P450 family 17 subfamily A member 1
|
|
chr12_-_119804472
Show fit
|
0.05 |
ENST00000678087.1
ENST00000677993.1
|
CIT
|
citron rho-interacting serine/threonine kinase
|
|
chr9_+_34990250
Show fit
|
0.05 |
ENST00000454002.6
ENST00000545841.5
|
DNAJB5
|
DnaJ heat shock protein family (Hsp40) member B5
|
|
chr9_+_121074944
Show fit
|
0.05 |
ENST00000373855.7
|
CNTRL
|
centriolin
|
|
chr1_+_161766309
Show fit
|
0.05 |
ENST00000679853.1
ENST00000681492.1
ENST00000679886.1
ENST00000367942.4
ENST00000680462.1
ENST00000680633.1
ENST00000681912.1
|
ATF6
|
activating transcription factor 6
|
|
chr12_-_119804298
Show fit
|
0.05 |
ENST00000678652.1
ENST00000678494.1
|
CIT
|
citron rho-interacting serine/threonine kinase
|
|
chr16_+_56336727
Show fit
|
0.05 |
ENST00000568375.2
|
GNAO1
|
G protein subunit alpha o1
|
|
chr13_-_38990824
Show fit
|
0.05 |
ENST00000379631.9
|
STOML3
|
stomatin like 3
|
|
chr7_-_29195186
Show fit
|
0.05 |
ENST00000449801.5
ENST00000409850.5
|
CPVL
|
carboxypeptidase vitellogenic like
|
|
chr8_-_29263063
Show fit
|
0.05 |
ENST00000524189.6
|
KIF13B
|
kinesin family member 13B
|
|
chr16_+_69424610
Show fit
|
0.05 |
ENST00000307892.13
|
CYB5B
|
cytochrome b5 type B
|
|
chr17_-_44268119
Show fit
|
0.05 |
ENST00000399246.3
ENST00000262418.12
|
SLC4A1
|
solute carrier family 4 member 1 (Diego blood group)
|
|
chr14_-_106627685
Show fit
|
0.05 |
ENST00000390629.3
|
IGHV4-59
|
immunoglobulin heavy variable 4-59
|
|
chr7_-_150978284
Show fit
|
0.04 |
ENST00000262186.10
|
KCNH2
|
potassium voltage-gated channel subfamily H member 2
|
|
chr5_-_115169894
Show fit
|
0.04 |
ENST00000513154.6
ENST00000508894.1
|
TRIM36
|
tripartite motif containing 36
|
|
chr11_+_61680373
Show fit
|
0.04 |
ENST00000257215.10
|
DAGLA
|
diacylglycerol lipase alpha
|
|
chr16_-_970954
Show fit
|
0.04 |
ENST00000543238.5
ENST00000262301.16
|
LMF1
|
lipase maturation factor 1
|
|
chr11_-_111879154
Show fit
|
0.04 |
ENST00000260257.9
|
FDXACB1
|
ferredoxin-fold anticodon binding domain containing 1
|
|
chrX_-_13817027
Show fit
|
0.04 |
ENST00000493677.5
ENST00000355135.6
ENST00000316715.9
|
GPM6B
|
glycoprotein M6B
|
|
chr19_-_58098203
Show fit
|
0.04 |
ENST00000600845.1
ENST00000240727.10
ENST00000600897.5
ENST00000421612.6
ENST00000601063.1
ENST00000601144.6
|
ZSCAN18
|
zinc finger and SCAN domain containing 18
|
|
chr15_+_63121818
Show fit
|
0.04 |
ENST00000413507.3
|
LACTB
|
lactamase beta
|
|
chr16_-_1230089
Show fit
|
0.04 |
ENST00000612142.1
ENST00000606293.5
|
TPSB2
|
tryptase beta 2
|
|
chr17_-_27893339
Show fit
|
0.04 |
ENST00000460380.6
ENST00000379102.8
ENST00000508862.5
ENST00000582441.1
|
LYRM9
ENSG00000266202.1
|
LYR motif containing 9
novel protein
|
|
chr19_-_54339146
Show fit
|
0.04 |
ENST00000291759.5
|
LILRA4
|
leukocyte immunoglobulin like receptor A4
|
|
chr6_+_42916046
Show fit
|
0.04 |
ENST00000304672.6
|
PTCRA
|
pre T cell antigen receptor alpha
|
|
chr19_+_18419322
Show fit
|
0.04 |
ENST00000348495.10
|
SSBP4
|
single stranded DNA binding protein 4
|
|
chr11_+_104036624
Show fit
|
0.04 |
ENST00000302259.5
|
DDI1
|
DNA damage inducible 1 homolog 1
|
|
chr13_-_38990856
Show fit
|
0.04 |
ENST00000423210.1
|
STOML3
|
stomatin like 3
|
|
chr2_-_74391837
Show fit
|
0.04 |
ENST00000417090.1
ENST00000409868.5
ENST00000680606.1
|
DCTN1
|
dynactin subunit 1
|
|
chr19_-_51366338
Show fit
|
0.04 |
ENST00000596253.1
ENST00000309244.9
|
ETFB
|
electron transfer flavoprotein subunit beta
|
|
chr19_+_18419374
Show fit
|
0.04 |
ENST00000270061.12
|
SSBP4
|
single stranded DNA binding protein 4
|
|
chr15_-_51622738
Show fit
|
0.04 |
ENST00000560891.6
|
DMXL2
|
Dmx like 2
|
|
chr17_-_73092657
Show fit
|
0.04 |
ENST00000580557.5
ENST00000579732.5
ENST00000578620.1
ENST00000542342.6
ENST00000255559.7
ENST00000579018.5
|
SLC39A11
|
solute carrier family 39 member 11
|
|
chr19_+_1000419
Show fit
|
0.04 |
ENST00000234389.3
|
GRIN3B
|
glutamate ionotropic receptor NMDA type subunit 3B
|
|
chr6_-_111873421
Show fit
|
0.04 |
ENST00000368678.8
ENST00000523238.5
ENST00000354650.7
|
FYN
|
FYN proto-oncogene, Src family tyrosine kinase
|
|
chrX_-_153673678
Show fit
|
0.04 |
ENST00000370150.5
|
PNCK
|
pregnancy up-regulated nonubiquitous CaM kinase
|
|
chrX_-_153673778
Show fit
|
0.04 |
ENST00000340888.8
|
PNCK
|
pregnancy up-regulated nonubiquitous CaM kinase
|
|
chr17_-_63881842
Show fit
|
0.04 |
ENST00000449787.6
ENST00000456543.6
ENST00000622506.4
ENST00000332800.7
ENST00000423893.7
|
GH2
|
growth hormone 2
|
|
chr3_+_9649433
Show fit
|
0.04 |
ENST00000353332.9
ENST00000420925.5
ENST00000296003.9
ENST00000351233.9
|
MTMR14
|
myotubularin related protein 14
|
|
chr7_+_93232340
Show fit
|
0.04 |
ENST00000251739.9
ENST00000544910.5
ENST00000305866.10
ENST00000458530.1
|
VPS50
|
VPS50 subunit of EARP/GARPII complex
|
|
chr8_+_7926337
Show fit
|
0.03 |
ENST00000400120.3
|
ZNF705B
|
zinc finger protein 705B
|
|
chr6_+_34236865
Show fit
|
0.03 |
ENST00000674029.1
ENST00000447654.5
ENST00000347617.10
ENST00000401473.7
ENST00000311487.9
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr15_-_51622798
Show fit
|
0.03 |
ENST00000251076.9
|
DMXL2
|
Dmx like 2
|
|
chr11_+_124954175
Show fit
|
0.03 |
ENST00000344762.6
|
CCDC15
|
coiled-coil domain containing 15
|
|
chr1_-_2003808
Show fit
|
0.03 |
ENST00000493964.5
|
CFAP74
|
cilia and flagella associated protein 74
|
|
chr11_-_615921
Show fit
|
0.03 |
ENST00000348655.11
ENST00000525445.6
ENST00000330243.9
|
IRF7
|
interferon regulatory factor 7
|
|
chr2_+_219514477
Show fit
|
0.03 |
ENST00000347842.8
ENST00000358078.5
|
ASIC4
|
acid sensing ion channel subunit family member 4
|
|
chr5_+_71719267
Show fit
|
0.03 |
ENST00000296777.5
|
CARTPT
|
CART prepropeptide
|
|
chr1_+_168280872
Show fit
|
0.03 |
ENST00000367821.8
|
TBX19
|
T-box transcription factor 19
|
|
chr7_+_2558960
Show fit
|
0.03 |
ENST00000623361.3
ENST00000402050.7
|
IQCE
|
IQ motif containing E
|
|
chr17_+_35121609
Show fit
|
0.03 |
ENST00000158009.6
|
FNDC8
|
fibronectin type III domain containing 8
|
|
chr14_+_100323332
Show fit
|
0.03 |
ENST00000361529.5
ENST00000557052.1
|
SLC25A47
|
solute carrier family 25 member 47
|
|
chr1_-_20786610
Show fit
|
0.03 |
ENST00000375000.5
ENST00000312239.10
ENST00000419490.5
ENST00000414993.1
ENST00000443615.1
|
HP1BP3
|
heterochromatin protein 1 binding protein 3
|
|
chr8_+_66043413
Show fit
|
0.03 |
ENST00000522619.1
|
DNAJC5B
|
DnaJ heat shock protein family (Hsp40) member C5 beta
|
|
chr17_-_46979240
Show fit
|
0.03 |
ENST00000322329.5
|
RPRML
|
reprimo like
|
|
chr2_+_184598520
Show fit
|
0.03 |
ENST00000302277.7
|
ZNF804A
|
zinc finger protein 804A
|
|
chr15_-_73992272
Show fit
|
0.03 |
ENST00000541638.6
ENST00000564777.5
ENST00000566081.5
ENST00000316911.10
ENST00000316900.9
|
STOML1
|
stomatin like 1
|
|
chr16_-_69384747
Show fit
|
0.03 |
ENST00000566257.5
|
TERF2
|
telomeric repeat binding factor 2
|
|
chr5_-_16738341
Show fit
|
0.03 |
ENST00000515803.5
|
MYO10
|
myosin X
|
|
chr5_-_59586393
Show fit
|
0.03 |
ENST00000505453.1
ENST00000360047.9
|
PDE4D
|
phosphodiesterase 4D
|
|
chr10_+_87863595
Show fit
|
0.02 |
ENST00000371953.8
|
PTEN
|
phosphatase and tensin homolog
|
|
chr5_+_122775062
Show fit
|
0.02 |
ENST00000379516.7
ENST00000505934.5
ENST00000514949.1
|
SNX2
|
sorting nexin 2
|
|
chr6_-_30160880
Show fit
|
0.02 |
ENST00000376704.3
|
TRIM10
|
tripartite motif containing 10
|
|
chr5_-_131994225
Show fit
|
0.02 |
ENST00000543479.5
ENST00000431707.5
|
ACSL6
|
acyl-CoA synthetase long chain family member 6
|
|
chr5_-_131994579
Show fit
|
0.02 |
ENST00000379240.5
|
ACSL6
|
acyl-CoA synthetase long chain family member 6
|
|
chr2_-_74392025
Show fit
|
0.02 |
ENST00000440727.1
ENST00000409240.5
|
DCTN1
|
dynactin subunit 1
|
|
chr5_+_156326735
Show fit
|
0.02 |
ENST00000435422.7
|
SGCD
|
sarcoglycan delta
|
|
chrX_+_17737443
Show fit
|
0.02 |
ENST00000398080.5
ENST00000380045.7
ENST00000380043.7
ENST00000380041.8
|
SCML1
|
Scm polycomb group protein like 1
|
|
chr13_+_32586443
Show fit
|
0.02 |
ENST00000315596.15
|
PDS5B
|
PDS5 cohesin associated factor B
|
|
chr19_+_9827886
Show fit
|
0.02 |
ENST00000358666.7
ENST00000590068.1
ENST00000593087.1
ENST00000586895.6
|
UBL5
|
ubiquitin like 5
|
|
chr11_+_111878926
Show fit
|
0.02 |
ENST00000528125.5
|
C11orf1
|
chromosome 11 open reading frame 1
|
|
chr1_-_56819365
Show fit
|
0.02 |
ENST00000343433.7
|
FYB2
|
FYN binding protein 2
|
|
chr9_-_127778659
Show fit
|
0.02 |
ENST00000314830.13
|
SH2D3C
|
SH2 domain containing 3C
|
|
chr19_+_11814256
Show fit
|
0.02 |
ENST00000304060.10
ENST00000457526.1
ENST00000427505.5
|
ZNF440
|
zinc finger protein 440
|
|
chr10_-_100286341
Show fit
|
0.02 |
ENST00000441611.5
ENST00000614731.4
|
BLOC1S2
|
biogenesis of lysosomal organelles complex 1 subunit 2
|
|
chr8_-_142742398
Show fit
|
0.02 |
ENST00000246515.2
|
SLURP1
|
secreted LY6/PLAUR domain containing 1
|
|
chr2_-_197435002
Show fit
|
0.02 |
ENST00000335508.11
ENST00000414963.2
ENST00000487698.5
|
SF3B1
|
splicing factor 3b subunit 1
|
|
chr18_+_56651385
Show fit
|
0.02 |
ENST00000615645.4
|
WDR7
|
WD repeat domain 7
|
|
chr10_+_4826200
Show fit
|
0.02 |
ENST00000298375.12
|
AKR1E2
|
aldo-keto reductase family 1 member E2
|
|
chr5_+_112976757
Show fit
|
0.02 |
ENST00000389063.3
|
DCP2
|
decapping mRNA 2
|
|
chrX_-_13734575
Show fit
|
0.02 |
ENST00000519885.5
ENST00000458511.7
ENST00000380579.6
ENST00000683983.1
ENST00000683569.1
ENST00000359680.9
|
TRAPPC2
|
trafficking protein particle complex 2
|
|
chr4_-_82561972
Show fit
|
0.02 |
ENST00000454948.3
ENST00000449862.7
|
TMEM150C
|
transmembrane protein 150C
|
|
chr18_+_56651335
Show fit
|
0.02 |
ENST00000589935.1
ENST00000254442.8
ENST00000357574.7
|
WDR7
|
WD repeat domain 7
|
|
chr3_-_131026726
Show fit
|
0.02 |
ENST00000514044.5
ENST00000264992.8
|
ASTE1
|
asteroid homolog 1
|
|
chr16_+_69424634
Show fit
|
0.02 |
ENST00000515314.6
ENST00000561792.6
ENST00000568237.1
|
CYB5B
|
cytochrome b5 type B
|
|
chr17_-_41047267
Show fit
|
0.02 |
ENST00000542137.1
ENST00000391419.3
|
KRTAP2-1
|
keratin associated protein 2-1
|
|
chr22_+_38656627
Show fit
|
0.02 |
ENST00000411557.5
ENST00000396811.6
ENST00000216029.7
ENST00000416285.5
|
CBY1
|
chibby family member 1, beta catenin antagonist
|
|
chr8_-_97277890
Show fit
|
0.02 |
ENST00000322128.5
|
TSPYL5
|
TSPY like 5
|
|
chr21_-_42315336
Show fit
|
0.02 |
ENST00000398431.2
ENST00000518498.3
|
TFF3
|
trefoil factor 3
|
|
chr12_-_122730828
Show fit
|
0.02 |
ENST00000432564.3
|
HCAR1
|
hydroxycarboxylic acid receptor 1
|
|
chr8_+_42697339
Show fit
|
0.02 |
ENST00000289957.3
ENST00000534391.1
|
CHRNB3
|
cholinergic receptor nicotinic beta 3 subunit
|
|
chr1_-_22143088
Show fit
|
0.02 |
ENST00000290167.11
|
WNT4
|
Wnt family member 4
|
|
chr11_+_119067774
Show fit
|
0.01 |
ENST00000621676.5
ENST00000614944.4
|
VPS11
|
VPS11 core subunit of CORVET and HOPS complexes
|
|
chr8_+_105318428
Show fit
|
0.01 |
ENST00000407775.7
|
ZFPM2
|
zinc finger protein, FOG family member 2
|
|
chr2_+_206274643
Show fit
|
0.01 |
ENST00000374423.9
ENST00000649650.1
|
ZDBF2
|
zinc finger DBF-type containing 2
|
|
chr20_+_325536
Show fit
|
0.01 |
ENST00000342665.5
|
SOX12
|
SRY-box transcription factor 12
|
|
chr1_+_19596960
Show fit
|
0.01 |
ENST00000617872.4
ENST00000322753.7
ENST00000602662.1
|
MICOS10
NBL1
|
mitochondrial contact site and cristae organizing system subunit 10
NBL1, DAN family BMP antagonist
|
|
chr9_+_93263948
Show fit
|
0.01 |
ENST00000448251.5
|
WNK2
|
WNK lysine deficient protein kinase 2
|
|
chrX_+_47561172
Show fit
|
0.01 |
ENST00000377045.9
ENST00000377039.2
|
ARAF
|
A-Raf proto-oncogene, serine/threonine kinase
|