|
chr5_+_31639104
Show fit
|
0.36 |
ENST00000438447.2
|
PDZD2
|
PDZ domain containing 2
|
|
chr12_-_57767057
Show fit
|
0.32 |
ENST00000228606.9
|
CYP27B1
|
cytochrome P450 family 27 subfamily B member 1
|
|
chr12_-_27971970
Show fit
|
0.23 |
ENST00000395872.5
ENST00000201015.8
|
PTHLH
|
parathyroid hormone like hormone
|
|
chr1_-_186680411
Show fit
|
0.23 |
ENST00000367468.10
|
PTGS2
|
prostaglandin-endoperoxide synthase 2
|
|
chr6_+_130365958
Show fit
|
0.21 |
ENST00000296978.4
|
TMEM200A
|
transmembrane protein 200A
|
|
chr19_+_40717091
Show fit
|
0.19 |
ENST00000263370.3
|
ITPKC
|
inositol-trisphosphate 3-kinase C
|
|
chr6_-_155455830
Show fit
|
0.19 |
ENST00000159060.3
|
NOX3
|
NADPH oxidase 3
|
|
chr6_+_14117764
Show fit
|
0.19 |
ENST00000379153.4
|
CD83
|
CD83 molecule
|
|
chr19_+_45001430
Show fit
|
0.17 |
ENST00000625761.2
ENST00000505236.1
ENST00000221452.13
|
RELB
|
RELB proto-oncogene, NF-kB subunit
|
|
chr2_+_17540670
Show fit
|
0.17 |
ENST00000451533.5
ENST00000295156.9
|
VSNL1
|
visinin like 1
|
|
chr19_-_4338786
Show fit
|
0.16 |
ENST00000601482.1
ENST00000600324.5
ENST00000594605.6
|
STAP2
|
signal transducing adaptor family member 2
|
|
chr20_+_6767678
Show fit
|
0.16 |
ENST00000378827.5
|
BMP2
|
bone morphogenetic protein 2
|
|
chr2_-_162318475
Show fit
|
0.16 |
ENST00000648433.1
|
IFIH1
|
interferon induced with helicase C domain 1
|
|
chr11_+_102317492
Show fit
|
0.16 |
ENST00000673846.1
|
BIRC3
|
baculoviral IAP repeat containing 3
|
|
chr11_+_34621065
Show fit
|
0.16 |
ENST00000257831.8
|
EHF
|
ETS homologous factor
|
|
chr16_+_67431112
Show fit
|
0.15 |
ENST00000326152.6
|
HSD11B2
|
hydroxysteroid 11-beta dehydrogenase 2
|
|
chr6_+_30882914
Show fit
|
0.14 |
ENST00000509639.5
ENST00000412274.6
ENST00000507901.5
ENST00000507046.5
ENST00000437124.6
ENST00000454612.6
ENST00000396342.6
|
DDR1
|
discoidin domain receptor tyrosine kinase 1
|
|
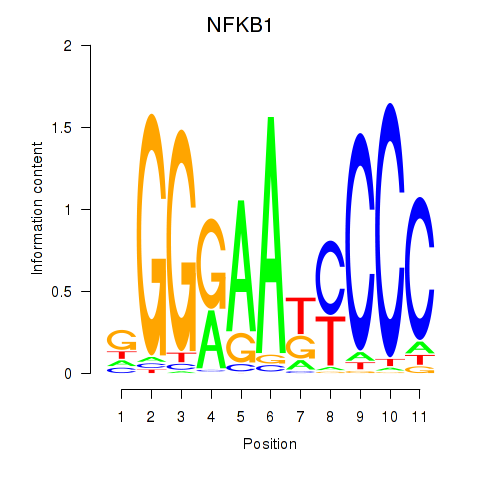
chr4_+_102501298
Show fit
|
0.14 |
ENST00000394820.8
ENST00000226574.9
ENST00000511926.5
ENST00000507079.5
|
NFKB1
|
nuclear factor kappa B subunit 1
|
|
chr11_+_1839452
Show fit
|
0.14 |
ENST00000381906.5
|
TNNI2
|
troponin I2, fast skeletal type
|
|
chr8_+_103819244
Show fit
|
0.14 |
ENST00000262231.14
ENST00000507740.5
ENST00000408894.6
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr11_+_1839602
Show fit
|
0.13 |
ENST00000617947.4
ENST00000252898.11
|
TNNI2
|
troponin I2, fast skeletal type
|
|
chr2_+_162318884
Show fit
|
0.13 |
ENST00000446271.5
ENST00000429691.6
|
GCA
|
grancalcin
|
|
chr17_+_9021501
Show fit
|
0.13 |
ENST00000173229.7
|
NTN1
|
netrin 1
|
|
chr1_-_209651291
Show fit
|
0.12 |
ENST00000391911.5
ENST00000415782.1
|
LAMB3
|
laminin subunit beta 3
|
|
chr10_+_102394488
Show fit
|
0.11 |
ENST00000369966.8
|
NFKB2
|
nuclear factor kappa B subunit 2
|
|
chr6_+_137867241
Show fit
|
0.11 |
ENST00000612899.5
ENST00000420009.5
|
TNFAIP3
|
TNF alpha induced protein 3
|
|
chr6_+_137867414
Show fit
|
0.11 |
ENST00000237289.8
ENST00000433680.1
|
TNFAIP3
|
TNF alpha induced protein 3
|
|
chr19_+_1067144
Show fit
|
0.11 |
ENST00000313093.7
|
ARHGAP45
|
Rho GTPase activating protein 45
|
|
chr19_+_1067493
Show fit
|
0.11 |
ENST00000586866.5
|
ARHGAP45
|
Rho GTPase activating protein 45
|
|
chr1_+_61952036
Show fit
|
0.11 |
ENST00000646453.1
ENST00000635137.1
|
PATJ
|
PATJ crumbs cell polarity complex component
|
|
chr1_+_61952283
Show fit
|
0.11 |
ENST00000307297.8
|
PATJ
|
PATJ crumbs cell polarity complex component
|
|
chr17_-_7294592
Show fit
|
0.11 |
ENST00000007699.10
|
YBX2
|
Y-box binding protein 2
|
|
chr10_-_71773513
Show fit
|
0.10 |
ENST00000394957.8
|
VSIR
|
V-set immunoregulatory receptor
|
|
chr11_+_34621109
Show fit
|
0.10 |
ENST00000450654.6
|
EHF
|
ETS homologous factor
|
|
chr9_+_17579059
Show fit
|
0.10 |
ENST00000380607.5
|
SH3GL2
|
SH3 domain containing GRB2 like 2, endophilin A1
|
|
chr19_+_1067272
Show fit
|
0.10 |
ENST00000590214.5
|
ARHGAP45
|
Rho GTPase activating protein 45
|
|
chr16_-_57284654
Show fit
|
0.10 |
ENST00000613167.4
ENST00000219207.10
ENST00000569059.5
|
PLLP
|
plasmolipin
|
|
chr2_+_64454506
Show fit
|
0.10 |
ENST00000409537.2
|
LGALSL
|
galectin like
|
|
chr8_-_138914034
Show fit
|
0.09 |
ENST00000303045.11
|
COL22A1
|
collagen type XXII alpha 1 chain
|
|
chr20_+_9514562
Show fit
|
0.09 |
ENST00000246070.3
|
LAMP5
|
lysosomal associated membrane protein family member 5
|
|
chr14_-_35404650
Show fit
|
0.09 |
ENST00000553342.1
ENST00000557140.5
ENST00000216797.10
|
NFKBIA
|
NFKB inhibitor alpha
|
|
chr6_+_106086316
Show fit
|
0.09 |
ENST00000369091.6
ENST00000369096.9
|
PRDM1
|
PR/SET domain 1
|
|
chr12_+_72272360
Show fit
|
0.08 |
ENST00000547300.2
ENST00000261180.10
|
TRHDE
|
thyrotropin releasing hormone degrading enzyme
|
|
chr19_-_38256339
Show fit
|
0.08 |
ENST00000591291.5
ENST00000301242.9
|
PPP1R14A
|
protein phosphatase 1 regulatory inhibitor subunit 14A
|
|
chr3_-_112133218
Show fit
|
0.08 |
ENST00000488580.5
ENST00000308910.9
ENST00000460387.6
ENST00000484193.5
ENST00000487901.1
|
GCSAM
|
germinal center associated signaling and motility
|
|
chr11_-_58578096
Show fit
|
0.08 |
ENST00000528954.5
ENST00000528489.1
|
LPXN
|
leupaxin
|
|
chr4_-_76023489
Show fit
|
0.08 |
ENST00000306602.3
|
CXCL10
|
C-X-C motif chemokine ligand 10
|
|
chr9_-_114806031
Show fit
|
0.08 |
ENST00000374045.5
|
TNFSF15
|
TNF superfamily member 15
|
|
chr19_-_38256513
Show fit
|
0.08 |
ENST00000347262.8
ENST00000591585.1
|
PPP1R14A
|
protein phosphatase 1 regulatory inhibitor subunit 14A
|
|
chr1_+_111227699
Show fit
|
0.07 |
ENST00000369748.9
|
CHI3L2
|
chitinase 3 like 2
|
|
chr1_+_111227610
Show fit
|
0.07 |
ENST00000369744.6
|
CHI3L2
|
chitinase 3 like 2
|
|
chr18_-_31162849
Show fit
|
0.07 |
ENST00000257197.7
ENST00000257198.6
|
DSC1
|
desmocollin 1
|
|
chr1_-_24143112
Show fit
|
0.07 |
ENST00000270800.2
|
IL22RA1
|
interleukin 22 receptor subunit alpha 1
|
|
chr2_-_74441882
Show fit
|
0.07 |
ENST00000272430.10
|
RTKN
|
rhotekin
|
|
chr13_+_97222296
Show fit
|
0.07 |
ENST00000343600.8
ENST00000376673.7
ENST00000679496.1
ENST00000345429.10
|
MBNL2
|
muscleblind like splicing regulator 2
|
|
chr11_+_102317542
Show fit
|
0.06 |
ENST00000532808.5
|
BIRC3
|
baculoviral IAP repeat containing 3
|
|
chr15_-_88895511
Show fit
|
0.06 |
ENST00000562281.1
ENST00000359595.8
ENST00000562889.5
|
HAPLN3
|
hyaluronan and proteoglycan link protein 3
|
|
chr6_+_12012304
Show fit
|
0.06 |
ENST00000379388.7
ENST00000627968.2
ENST00000541134.5
|
HIVEP1
|
HIVEP zinc finger 1
|
|
chr19_-_4831689
Show fit
|
0.06 |
ENST00000248244.6
|
TICAM1
|
toll like receptor adaptor molecule 1
|
|
chr4_+_73869385
Show fit
|
0.06 |
ENST00000395761.4
|
CXCL1
|
C-X-C motif chemokine ligand 1
|
|
chr11_+_102317450
Show fit
|
0.06 |
ENST00000615299.4
ENST00000527309.2
ENST00000526421.6
ENST00000263464.9
|
BIRC3
|
baculoviral IAP repeat containing 3
|
|
chrX_+_153411472
Show fit
|
0.06 |
ENST00000338647.7
|
ZFP92
|
ZFP92 zinc finger protein
|
|
chr2_+_112977998
Show fit
|
0.06 |
ENST00000259205.5
ENST00000376489.6
|
IL36G
|
interleukin 36 gamma
|
|
chr1_-_205775449
Show fit
|
0.06 |
ENST00000235932.8
ENST00000437324.6
ENST00000414729.1
ENST00000367139.8
|
RAB29
|
RAB29, member RAS oncogene family
|
|
chr1_+_156153568
Show fit
|
0.06 |
ENST00000368284.5
ENST00000368286.6
ENST00000368285.8
ENST00000438830.5
|
SEMA4A
|
semaphorin 4A
|
|
chr15_+_85380625
Show fit
|
0.06 |
ENST00000560302.5
|
AKAP13
|
A-kinase anchoring protein 13
|
|
chr3_+_50269140
Show fit
|
0.06 |
ENST00000616701.5
ENST00000433753.4
ENST00000611067.4
|
SEMA3B
|
semaphorin 3B
|
|
chr2_-_100104530
Show fit
|
0.06 |
ENST00000432037.5
ENST00000673232.1
ENST00000423966.6
ENST00000409236.6
|
AFF3
|
AF4/FMR2 family member 3
|
|
chr12_+_56338873
Show fit
|
0.05 |
ENST00000228534.6
|
IL23A
|
interleukin 23 subunit alpha
|
|
chr6_+_147508645
Show fit
|
0.05 |
ENST00000367474.2
|
SAMD5
|
sterile alpha motif domain containing 5
|
|
chr2_+_60881515
Show fit
|
0.05 |
ENST00000295025.12
|
REL
|
REL proto-oncogene, NF-kB subunit
|
|
chr15_-_74725370
Show fit
|
0.05 |
ENST00000567032.5
ENST00000564596.5
ENST00000566503.1
ENST00000395049.8
ENST00000379727.8
ENST00000617691.4
ENST00000395048.6
|
CYP1A1
|
cytochrome P450 family 1 subfamily A member 1
|
|
chr1_-_10796636
Show fit
|
0.05 |
ENST00000377022.8
ENST00000344008.5
|
CASZ1
|
castor zinc finger 1
|
|
chr2_-_162318613
Show fit
|
0.05 |
ENST00000649979.2
ENST00000421365.2
|
IFIH1
|
interferon induced with helicase C domain 1
|
|
chr12_+_48106094
Show fit
|
0.05 |
ENST00000546755.5
ENST00000549366.5
ENST00000642730.1
ENST00000552792.5
|
PFKM
|
phosphofructokinase, muscle
|
|
chr1_-_205775182
Show fit
|
0.05 |
ENST00000446390.6
|
RAB29
|
RAB29, member RAS oncogene family
|
|
chr4_-_184474518
Show fit
|
0.05 |
ENST00000393593.8
|
IRF2
|
interferon regulatory factor 2
|
|
chr1_+_37474572
Show fit
|
0.05 |
ENST00000373087.7
|
ZC3H12A
|
zinc finger CCCH-type containing 12A
|
|
chr2_+_60881553
Show fit
|
0.05 |
ENST00000394479.4
|
REL
|
REL proto-oncogene, NF-kB subunit
|
|
chr3_+_6861107
Show fit
|
0.05 |
ENST00000357716.9
ENST00000486284.5
ENST00000389336.8
|
GRM7
|
glutamate metabotropic receptor 7
|
|
chr6_+_135181323
Show fit
|
0.05 |
ENST00000367814.8
|
MYB
|
MYB proto-oncogene, transcription factor
|
|
chr12_-_26125023
Show fit
|
0.05 |
ENST00000242728.5
|
BHLHE41
|
basic helix-loop-helix family member e41
|
|
chr14_+_32939243
Show fit
|
0.05 |
ENST00000346562.6
ENST00000548645.5
ENST00000356141.8
ENST00000357798.9
|
NPAS3
|
neuronal PAS domain protein 3
|
|
chr1_-_155990062
Show fit
|
0.04 |
ENST00000462460.6
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor 2
|
|
chr14_+_75069577
Show fit
|
0.04 |
ENST00000238686.8
|
ZC2HC1C
|
zinc finger C2HC-type containing 1C
|
|
chrX_-_15854743
Show fit
|
0.04 |
ENST00000450644.2
|
AP1S2
|
adaptor related protein complex 1 subunit sigma 2
|
|
chr12_-_54981838
Show fit
|
0.04 |
ENST00000316577.12
|
TESPA1
|
thymocyte expressed, positive selection associated 1
|
|
chr6_+_148747016
Show fit
|
0.04 |
ENST00000367463.5
|
UST
|
uronyl 2-sulfotransferase
|
|
chrX_-_80809854
Show fit
|
0.04 |
ENST00000373275.5
|
BRWD3
|
bromodomain and WD repeat domain containing 3
|
|
chrX_-_112840815
Show fit
|
0.04 |
ENST00000304758.5
ENST00000371959.9
|
AMOT
|
angiomotin
|
|
chr12_+_52051402
Show fit
|
0.04 |
ENST00000243050.5
ENST00000550763.1
ENST00000394825.6
ENST00000394824.2
ENST00000548232.1
ENST00000562373.1
|
NR4A1
|
nuclear receptor subfamily 4 group A member 1
|
|
chrX_-_20266606
Show fit
|
0.04 |
ENST00000643085.1
ENST00000645270.1
ENST00000644368.1
|
RPS6KA3
|
ribosomal protein S6 kinase A3
|
|
chr16_+_50696999
Show fit
|
0.04 |
ENST00000300589.6
|
NOD2
|
nucleotide binding oligomerization domain containing 2
|
|
chr17_-_42181116
Show fit
|
0.04 |
ENST00000264661.4
|
KCNH4
|
potassium voltage-gated channel subfamily H member 4
|
|
chr6_+_32154131
Show fit
|
0.04 |
ENST00000375143.6
ENST00000324816.11
ENST00000424499.1
|
PPT2
|
palmitoyl-protein thioesterase 2
|
|
chr19_-_45730861
Show fit
|
0.04 |
ENST00000317683.4
|
FBXO46
|
F-box protein 46
|
|
chr17_+_42567072
Show fit
|
0.04 |
ENST00000246912.8
ENST00000346833.8
ENST00000591024.1
|
MLX
|
MAX dimerization protein MLX
|
|
chr6_+_135181268
Show fit
|
0.04 |
ENST00000341911.10
ENST00000442647.7
ENST00000618728.4
ENST00000316528.12
ENST00000616088.4
|
MYB
|
MYB proto-oncogene, transcription factor
|
|
chr14_+_23372809
Show fit
|
0.04 |
ENST00000397242.2
ENST00000329715.2
|
IL25
|
interleukin 25
|
|
chr8_-_144358458
Show fit
|
0.04 |
ENST00000331890.6
ENST00000455319.6
|
FBXL6
|
F-box and leucine rich repeat protein 6
|
|
chr17_-_42181081
Show fit
|
0.04 |
ENST00000607371.5
|
KCNH4
|
potassium voltage-gated channel subfamily H member 4
|
|
chr8_+_22042382
Show fit
|
0.04 |
ENST00000518533.5
|
FGF17
|
fibroblast growth factor 17
|
|
chr3_-_66038537
Show fit
|
0.04 |
ENST00000483466.5
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1
|
|
chr22_-_37984534
Show fit
|
0.04 |
ENST00000396884.8
|
SOX10
|
SRY-box transcription factor 10
|
|
chr6_+_29723340
Show fit
|
0.04 |
ENST00000334668.8
|
HLA-F
|
major histocompatibility complex, class I, F
|
|
chr1_-_205935822
Show fit
|
0.04 |
ENST00000340781.8
|
SLC26A9
|
solute carrier family 26 member 9
|
|
chr1_-_36149450
Show fit
|
0.04 |
ENST00000373163.5
|
TRAPPC3
|
trafficking protein particle complex 3
|
|
chr7_-_111562455
Show fit
|
0.04 |
ENST00000452895.5
ENST00000405709.7
ENST00000452753.1
ENST00000331762.7
|
IMMP2L
|
inner mitochondrial membrane peptidase subunit 2
|
|
chr21_+_33403391
Show fit
|
0.04 |
ENST00000290219.11
ENST00000381995.5
|
IFNGR2
|
interferon gamma receptor 2
|
|
chr17_+_7351889
Show fit
|
0.04 |
ENST00000576980.2
|
KCTD11
|
potassium channel tetramerization domain containing 11
|
|
chr6_-_31582415
Show fit
|
0.04 |
ENST00000429299.3
ENST00000446745.2
|
LTB
|
lymphotoxin beta
|
|
chr14_+_75069632
Show fit
|
0.04 |
ENST00000439583.2
ENST00000554763.2
ENST00000524913.3
ENST00000525046.2
ENST00000674086.1
ENST00000526130.2
ENST00000674094.1
ENST00000532198.2
|
ZC2HC1C
|
zinc finger C2HC-type containing 1C
|
|
chr14_+_102777461
Show fit
|
0.04 |
ENST00000560371.5
ENST00000347662.8
|
TRAF3
|
TNF receptor associated factor 3
|
|
chr6_+_29723421
Show fit
|
0.03 |
ENST00000259951.12
ENST00000434407.6
|
HLA-F
|
major histocompatibility complex, class I, F
|
|
chr19_-_2042066
Show fit
|
0.03 |
ENST00000591588.1
ENST00000591142.5
|
MKNK2
|
MAPK interacting serine/threonine kinase 2
|
|
chr17_+_42567084
Show fit
|
0.03 |
ENST00000435881.7
|
MLX
|
MAX dimerization protein MLX
|
|
chr6_+_32154010
Show fit
|
0.03 |
ENST00000375137.6
|
PPT2
|
palmitoyl-protein thioesterase 2
|
|
chr8_-_102655707
Show fit
|
0.03 |
ENST00000285407.11
|
KLF10
|
Kruppel like factor 10
|
|
chrX_+_148500883
Show fit
|
0.03 |
ENST00000370458.5
ENST00000370457.9
|
AFF2
|
AF4/FMR2 family member 2
|
|
chr5_-_79069668
Show fit
|
0.03 |
ENST00000255189.8
|
DMGDH
|
dimethylglycine dehydrogenase
|
|
chr7_-_56034133
Show fit
|
0.03 |
ENST00000421626.5
|
PSPH
|
phosphoserine phosphatase
|
|
chr15_+_73052449
Show fit
|
0.03 |
ENST00000261908.11
|
NEO1
|
neogenin 1
|
|
chr1_-_36149464
Show fit
|
0.03 |
ENST00000373166.8
ENST00000373159.1
ENST00000373162.5
ENST00000616074.4
ENST00000616395.4
|
TRAPPC3
|
trafficking protein particle complex 3
|
|
chr1_+_6785437
Show fit
|
0.03 |
ENST00000303635.12
ENST00000473578.5
ENST00000557126.5
|
CAMTA1
|
calmodulin binding transcription activator 1
|
|
chr11_-_35419213
Show fit
|
0.03 |
ENST00000642171.1
ENST00000644050.1
ENST00000643134.1
|
SLC1A2
|
solute carrier family 1 member 2
|
|
chr13_+_76992412
Show fit
|
0.03 |
ENST00000636705.1
|
CLN5
|
CLN5 intracellular trafficking protein
|
|
chr19_-_48646155
Show fit
|
0.03 |
ENST00000084798.9
|
CA11
|
carbonic anhydrase 11
|
|
chr12_+_48105466
Show fit
|
0.03 |
ENST00000549003.5
ENST00000550924.6
|
PFKM
|
phosphofructokinase, muscle
|
|
chr19_+_49591170
Show fit
|
0.03 |
ENST00000418929.7
|
PRR12
|
proline rich 12
|
|
chr21_+_33403466
Show fit
|
0.03 |
ENST00000405436.5
|
IFNGR2
|
interferon gamma receptor 2
|
|
chr7_-_27200083
Show fit
|
0.03 |
ENST00000649031.1
|
HOXA13
|
homeobox A13
|
|
chr10_-_124092445
Show fit
|
0.03 |
ENST00000346248.7
|
CHST15
|
carbohydrate sulfotransferase 15
|
|
chr16_+_6019585
Show fit
|
0.03 |
ENST00000547372.5
|
RBFOX1
|
RNA binding fox-1 homolog 1
|
|
chr17_+_21288029
Show fit
|
0.03 |
ENST00000526076.6
ENST00000361818.9
ENST00000316920.10
|
MAP2K3
|
mitogen-activated protein kinase kinase 3
|
|
chr11_-_35419098
Show fit
|
0.03 |
ENST00000606205.6
ENST00000645303.1
|
SLC1A2
|
solute carrier family 1 member 2
|
|
chr19_+_44748673
Show fit
|
0.03 |
ENST00000164227.10
|
BCL3
|
BCL3 transcription coactivator
|
|
chr12_-_57129001
Show fit
|
0.03 |
ENST00000556155.5
|
STAT6
|
signal transducer and activator of transcription 6
|
|
chr3_-_187737943
Show fit
|
0.03 |
ENST00000438077.1
|
BCL6
|
BCL6 transcription repressor
|
|
chr17_+_7352142
Show fit
|
0.03 |
ENST00000333751.8
|
KCTD11
|
potassium channel tetramerization domain containing 11
|
|
chr4_-_40629842
Show fit
|
0.03 |
ENST00000295971.12
|
RBM47
|
RNA binding motif protein 47
|
|
chr1_-_1421302
Show fit
|
0.03 |
ENST00000520296.5
|
ANKRD65
|
ankyrin repeat domain 65
|
|
chr1_+_155687946
Show fit
|
0.03 |
ENST00000471642.6
ENST00000471214.5
|
DAP3
|
death associated protein 3
|
|
chr14_+_95876762
Show fit
|
0.03 |
ENST00000503525.2
|
TUNAR
|
TCL1 upstream neural differentiation-associated RNA
|
|
chr1_+_67166448
Show fit
|
0.03 |
ENST00000347310.10
|
IL23R
|
interleukin 23 receptor
|
|
chr3_+_23805941
Show fit
|
0.03 |
ENST00000306627.8
ENST00000346855.7
|
UBE2E1
|
ubiquitin conjugating enzyme E2 E1
|
|
chr19_+_35449584
Show fit
|
0.03 |
ENST00000246549.2
|
FFAR2
|
free fatty acid receptor 2
|
|
chr11_-_35418966
Show fit
|
0.03 |
ENST00000531628.2
|
SLC1A2
|
solute carrier family 1 member 2
|
|
chr1_+_107141022
Show fit
|
0.03 |
ENST00000370067.5
ENST00000370068.6
|
NTNG1
|
netrin G1
|
|
chr7_-_1556194
Show fit
|
0.03 |
ENST00000297477.10
|
TMEM184A
|
transmembrane protein 184A
|
|
chr3_+_9397602
Show fit
|
0.03 |
ENST00000402198.7
|
SETD5
|
SET domain containing 5
|
|
chr1_-_57424014
Show fit
|
0.03 |
ENST00000371230.1
ENST00000371236.7
|
DAB1
|
DAB adaptor protein 1
|
|
chr11_+_73289403
Show fit
|
0.03 |
ENST00000535931.2
ENST00000544437.6
|
P2RY6
|
pyrimidinergic receptor P2Y6
|
|
chr2_+_68467544
Show fit
|
0.03 |
ENST00000303795.9
|
APLF
|
aprataxin and PNKP like factor
|
|
chr14_+_75522427
Show fit
|
0.03 |
ENST00000286639.8
|
BATF
|
basic leucine zipper ATF-like transcription factor
|
|
chr14_+_102777555
Show fit
|
0.03 |
ENST00000539721.5
ENST00000560463.5
|
TRAF3
|
TNF receptor associated factor 3
|
|
chr12_+_57229694
Show fit
|
0.03 |
ENST00000557487.5
ENST00000328923.8
ENST00000555634.5
ENST00000556689.5
|
SHMT2
|
serine hydroxymethyltransferase 2
|
|
chr17_+_12665882
Show fit
|
0.03 |
ENST00000425538.6
|
MYOCD
|
myocardin
|
|
chr7_-_101237827
Show fit
|
0.03 |
ENST00000611078.4
|
CLDN15
|
claudin 15
|
|
chr3_+_53161120
Show fit
|
0.03 |
ENST00000394729.6
ENST00000330452.8
ENST00000652449.1
|
PRKCD
|
protein kinase C delta
|
|
chr6_-_32154326
Show fit
|
0.03 |
ENST00000475826.1
ENST00000485392.5
ENST00000494332.5
ENST00000498575.1
ENST00000428778.5
|
ENSG00000284954.1
ENSG00000285085.1
|
novel transcript
novel protein
|
|
chr14_+_102777433
Show fit
|
0.03 |
ENST00000392745.8
|
TRAF3
|
TNF receptor associated factor 3
|
|
chr6_-_142946312
Show fit
|
0.03 |
ENST00000367604.6
|
HIVEP2
|
HIVEP zinc finger 2
|
|
chr4_-_145938422
Show fit
|
0.03 |
ENST00000656985.1
ENST00000652097.1
ENST00000503462.3
ENST00000379448.9
ENST00000513840.2
|
ZNF827
|
zinc finger protein 827
|
|
chr14_+_103123452
Show fit
|
0.03 |
ENST00000558056.1
ENST00000560869.6
|
TNFAIP2
|
TNF alpha induced protein 2
|
|
chr17_+_17782108
Show fit
|
0.03 |
ENST00000395774.1
|
RAI1
|
retinoic acid induced 1
|
|
chr19_+_38899946
Show fit
|
0.03 |
ENST00000572515.5
ENST00000313582.6
ENST00000575359.5
|
NFKBIB
|
NFKB inhibitor beta
|
|
chr19_-_33064872
Show fit
|
0.03 |
ENST00000254260.8
|
RHPN2
|
rhophilin Rho GTPase binding protein 2
|
|
chr16_-_66925526
Show fit
|
0.03 |
ENST00000299759.11
ENST00000420652.5
|
RRAD
|
RRAD, Ras related glycolysis inhibitor and calcium channel regulator
|
|
chr19_-_40716869
Show fit
|
0.02 |
ENST00000677018.1
ENST00000324464.8
ENST00000594720.6
ENST00000677496.1
|
COQ8B
|
coenzyme Q8B
|
|
chr19_-_48321857
Show fit
|
0.02 |
ENST00000474199.6
|
ODAD1
|
outer dynein arm docking complex subunit 1
|
|
chr4_-_145938473
Show fit
|
0.02 |
ENST00000513320.5
|
ZNF827
|
zinc finger protein 827
|
|
chr10_-_13099652
Show fit
|
0.02 |
ENST00000378839.1
|
CCDC3
|
coiled-coil domain containing 3
|
|
chr4_+_113292925
Show fit
|
0.02 |
ENST00000673353.1
ENST00000505342.6
ENST00000672915.1
ENST00000509550.5
|
ANK2
|
ankyrin 2
|
|
chr6_-_27312258
Show fit
|
0.02 |
ENST00000444565.2
|
POM121L2
|
POM121 transmembrane nucleoporin like 2
|
|
chr13_-_52848632
Show fit
|
0.02 |
ENST00000377942.7
ENST00000338862.5
|
PCDH8
|
protocadherin 8
|
|
chrX_+_73447042
Show fit
|
0.02 |
ENST00000373514.3
|
CDX4
|
caudal type homeobox 4
|
|
chr4_+_113292838
Show fit
|
0.02 |
ENST00000672411.1
ENST00000673231.1
|
ANK2
|
ankyrin 2
|
|
chr5_-_150412743
Show fit
|
0.02 |
ENST00000353334.11
ENST00000009530.12
ENST00000377795.7
|
CD74
|
CD74 molecule
|
|
chr12_-_57110284
Show fit
|
0.02 |
ENST00000543873.6
ENST00000554663.5
ENST00000557635.5
|
STAT6
|
signal transducer and activator of transcription 6
|
|
chr7_-_101237791
Show fit
|
0.02 |
ENST00000308344.10
|
CLDN15
|
claudin 15
|
|
chr11_-_14892217
Show fit
|
0.02 |
ENST00000334636.10
|
CYP2R1
|
cytochrome P450 family 2 subfamily R member 1
|
|
chr17_-_7251691
Show fit
|
0.02 |
ENST00000574322.6
|
CTDNEP1
|
CTD nuclear envelope phosphatase 1
|
|
chr2_+_38666059
Show fit
|
0.02 |
ENST00000272252.10
ENST00000410063.5
|
GALM
|
galactose mutarotase
|
|
chr20_-_49915509
Show fit
|
0.02 |
ENST00000289431.10
|
SPATA2
|
spermatogenesis associated 2
|
|
chr19_+_4229502
Show fit
|
0.02 |
ENST00000221847.6
|
EBI3
|
Epstein-Barr virus induced 3
|
|
chr10_+_110497898
Show fit
|
0.02 |
ENST00000369583.4
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr2_+_218880844
Show fit
|
0.02 |
ENST00000258411.8
|
WNT10A
|
Wnt family member 10A
|
|
chr10_+_115093404
Show fit
|
0.02 |
ENST00000527407.5
|
ATRNL1
|
attractin like 1
|
|
chr17_-_41811930
Show fit
|
0.02 |
ENST00000393928.6
|
P3H4
|
prolyl 3-hydroxylase family member 4 (inactive)
|
|
chrX_+_151912458
Show fit
|
0.02 |
ENST00000431963.6
ENST00000682265.1
|
MAGEA4
|
MAGE family member A4
|
|
chr3_+_53161241
Show fit
|
0.02 |
ENST00000477794.2
ENST00000650739.1
|
PRKCD
|
protein kinase C delta
|
|
chr1_-_235649734
Show fit
|
0.02 |
ENST00000391854.7
|
GNG4
|
G protein subunit gamma 4
|
|
chr2_+_20667136
Show fit
|
0.02 |
ENST00000272224.5
|
GDF7
|
growth differentiation factor 7
|
|
chr17_+_62458641
Show fit
|
0.02 |
ENST00000582809.5
|
TLK2
|
tousled like kinase 2
|
|
chr8_-_119832826
Show fit
|
0.02 |
ENST00000378164.7
|
TAF2
|
TATA-box binding protein associated factor 2
|
|
chr2_+_201182873
Show fit
|
0.02 |
ENST00000360132.7
|
CASP10
|
caspase 10
|
|
chr17_+_76385256
Show fit
|
0.02 |
ENST00000392496.3
|
SPHK1
|
sphingosine kinase 1
|
|
chrX_+_69504320
Show fit
|
0.02 |
ENST00000252338.5
|
FAM155B
|
family with sequence similarity 155 member B
|
|
chr2_+_73984902
Show fit
|
0.02 |
ENST00000409262.8
|
TET3
|
tet methylcytosine dioxygenase 3
|
|
chr8_+_26291494
Show fit
|
0.02 |
ENST00000380737.8
ENST00000524169.5
|
PPP2R2A
|
protein phosphatase 2 regulatory subunit Balpha
|