|
chr1_-_209651291
Show fit
|
0.90 |
ENST00000391911.5
ENST00000415782.1
|
LAMB3
|
laminin subunit beta 3
|
|
chr18_+_23949847
Show fit
|
0.78 |
ENST00000588004.1
|
LAMA3
|
laminin subunit alpha 3
|
|
chr16_-_68236069
Show fit
|
0.66 |
ENST00000473183.7
ENST00000565858.5
|
ESRP2
|
epithelial splicing regulatory protein 2
|
|
chr12_-_52520371
Show fit
|
0.58 |
ENST00000549420.1
ENST00000252242.9
ENST00000551275.1
ENST00000546577.1
|
KRT5
|
keratin 5
|
|
chr12_-_57767057
Show fit
|
0.50 |
ENST00000228606.9
|
CYP27B1
|
cytochrome P450 family 27 subfamily B member 1
|
|
chr5_+_31639104
Show fit
|
0.50 |
ENST00000438447.2
|
PDZD2
|
PDZ domain containing 2
|
|
chr20_+_45174894
Show fit
|
0.46 |
ENST00000243924.4
|
PI3
|
peptidase inhibitor 3
|
|
chr8_+_85463997
Show fit
|
0.41 |
ENST00000285379.10
|
CA2
|
carbonic anhydrase 2
|
|
chr9_-_137054016
Show fit
|
0.36 |
ENST00000312665.7
ENST00000355097.7
|
ENTPD2
|
ectonucleoside triphosphate diphosphohydrolase 2
|
|
chr11_-_18248662
Show fit
|
0.36 |
ENST00000256733.9
|
SAA2
|
serum amyloid A2
|
|
chr8_-_144416481
Show fit
|
0.33 |
ENST00000276833.9
|
SLC39A4
|
solute carrier family 39 member 4
|
|
chr1_-_159945596
Show fit
|
0.31 |
ENST00000361509.7
ENST00000611023.1
ENST00000368094.6
|
IGSF9
|
immunoglobulin superfamily member 9
|
|
chr15_-_88895511
Show fit
|
0.31 |
ENST00000562281.1
ENST00000359595.8
ENST00000562889.5
|
HAPLN3
|
hyaluronan and proteoglycan link protein 3
|
|
chr11_-_18248632
Show fit
|
0.29 |
ENST00000524555.3
ENST00000528349.5
ENST00000526900.1
ENST00000529528.5
ENST00000414546.6
|
SAA2-SAA4
SAA2
|
SAA2-SAA4 readthrough
serum amyloid A2
|
|
chr1_-_186680411
Show fit
|
0.27 |
ENST00000367468.10
|
PTGS2
|
prostaglandin-endoperoxide synthase 2
|
|
chr3_+_111999189
Show fit
|
0.27 |
ENST00000455401.6
|
TAGLN3
|
transgelin 3
|
|
chr3_+_113947901
Show fit
|
0.27 |
ENST00000330212.7
ENST00000498275.5
|
ZDHHC23
|
zinc finger DHHC-type palmitoyltransferase 23
|
|
chr1_-_183590876
Show fit
|
0.27 |
ENST00000367536.5
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr3_+_111998915
Show fit
|
0.26 |
ENST00000478951.6
|
TAGLN3
|
transgelin 3
|
|
chr16_-_87869497
Show fit
|
0.26 |
ENST00000261622.5
|
SLC7A5
|
solute carrier family 7 member 5
|
|
chr3_+_111998739
Show fit
|
0.26 |
ENST00000393917.6
ENST00000273368.8
|
TAGLN3
|
transgelin 3
|
|
chr4_-_22516001
Show fit
|
0.25 |
ENST00000334304.10
|
ADGRA3
|
adhesion G protein-coupled receptor A3
|
|
chr3_+_113948004
Show fit
|
0.25 |
ENST00000638807.2
|
ZDHHC23
|
zinc finger DHHC-type palmitoyltransferase 23
|
|
chr16_+_57620077
Show fit
|
0.24 |
ENST00000567835.5
ENST00000569372.5
ENST00000563548.5
ENST00000562003.5
|
ADGRG1
|
adhesion G protein-coupled receptor G1
|
|
chr1_-_84997079
Show fit
|
0.24 |
ENST00000284027.5
ENST00000370608.8
|
MCOLN2
|
mucolipin TRP cation channel 2
|
|
chr17_+_9021501
Show fit
|
0.24 |
ENST00000173229.7
|
NTN1
|
netrin 1
|
|
chr16_+_57619942
Show fit
|
0.24 |
ENST00000568908.5
ENST00000568909.5
ENST00000566778.5
ENST00000561988.5
|
ADGRG1
|
adhesion G protein-coupled receptor G1
|
|
chr15_-_74725370
Show fit
|
0.24 |
ENST00000567032.5
ENST00000564596.5
ENST00000566503.1
ENST00000395049.8
ENST00000379727.8
ENST00000617691.4
ENST00000395048.6
|
CYP1A1
|
cytochrome P450 family 1 subfamily A member 1
|
|
chr3_+_111999326
Show fit
|
0.24 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3
|
|
chr12_+_56080155
Show fit
|
0.23 |
ENST00000267101.8
|
ERBB3
|
erb-b2 receptor tyrosine kinase 3
|
|
chr12_+_56079843
Show fit
|
0.22 |
ENST00000549282.5
ENST00000549061.5
ENST00000683059.1
|
ERBB3
|
erb-b2 receptor tyrosine kinase 3
|
|
chr6_+_14117764
Show fit
|
0.21 |
ENST00000379153.4
|
CD83
|
CD83 molecule
|
|
chr19_-_4338786
Show fit
|
0.21 |
ENST00000601482.1
ENST00000600324.5
ENST00000594605.6
|
STAP2
|
signal transducing adaptor family member 2
|
|
chr19_+_53867874
Show fit
|
0.20 |
ENST00000448420.5
ENST00000439000.5
ENST00000391771.1
ENST00000391770.9
|
MYADM
|
myeloid associated differentiation marker
|
|
chr1_-_24187246
Show fit
|
0.19 |
ENST00000374421.7
ENST00000374418.3
ENST00000327575.6
ENST00000327535.6
|
IFNLR1
|
interferon lambda receptor 1
|
|
chr11_+_60924452
Show fit
|
0.19 |
ENST00000453848.7
ENST00000544065.5
ENST00000005286.8
|
TMEM132A
|
transmembrane protein 132A
|
|
chr1_-_183590596
Show fit
|
0.19 |
ENST00000418089.5
ENST00000413720.5
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr6_+_18155399
Show fit
|
0.19 |
ENST00000650836.2
ENST00000449850.2
ENST00000297792.9
|
KDM1B
|
lysine demethylase 1B
|
|
chr8_-_65842051
Show fit
|
0.18 |
ENST00000401827.8
|
PDE7A
|
phosphodiesterase 7A
|
|
chr1_-_204494752
Show fit
|
0.17 |
ENST00000684373.1
|
PIK3C2B
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta
|
|
chrX_+_68829009
Show fit
|
0.16 |
ENST00000204961.5
|
EFNB1
|
ephrin B1
|
|
chr9_+_128149447
Show fit
|
0.16 |
ENST00000277480.7
ENST00000372998.1
|
LCN2
|
lipocalin 2
|
|
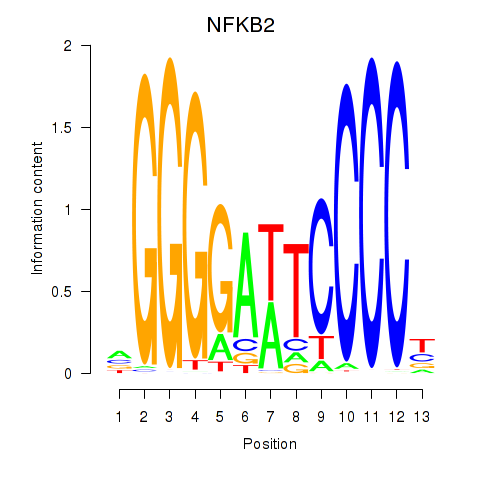
chr10_+_102394488
Show fit
|
0.16 |
ENST00000369966.8
|
NFKB2
|
nuclear factor kappa B subunit 2
|
|
chr4_+_73869385
Show fit
|
0.15 |
ENST00000395761.4
|
CXCL1
|
C-X-C motif chemokine ligand 1
|
|
chr19_+_45001430
Show fit
|
0.15 |
ENST00000625761.2
ENST00000505236.1
ENST00000221452.13
|
RELB
|
RELB proto-oncogene, NF-kB subunit
|
|
chr16_+_81444799
Show fit
|
0.15 |
ENST00000537098.8
|
CMIP
|
c-Maf inducing protein
|
|
chr11_+_67056805
Show fit
|
0.15 |
ENST00000308831.7
|
RHOD
|
ras homolog family member D
|
|
chr19_-_38256339
Show fit
|
0.15 |
ENST00000591291.5
ENST00000301242.9
|
PPP1R14A
|
protein phosphatase 1 regulatory inhibitor subunit 14A
|
|
chr1_-_6485433
Show fit
|
0.15 |
ENST00000535355.6
|
PLEKHG5
|
pleckstrin homology and RhoGEF domain containing G5
|
|
chr1_-_26360050
Show fit
|
0.15 |
ENST00000475866.3
|
CRYBG2
|
crystallin beta-gamma domain containing 2
|
|
chr19_-_38256513
Show fit
|
0.15 |
ENST00000347262.8
ENST00000591585.1
|
PPP1R14A
|
protein phosphatase 1 regulatory inhibitor subunit 14A
|
|
chr8_+_32548661
Show fit
|
0.14 |
ENST00000650980.1
ENST00000405005.7
|
NRG1
|
neuregulin 1
|
|
chr15_+_76995118
Show fit
|
0.14 |
ENST00000558012.6
ENST00000379595.7
|
PSTPIP1
|
proline-serine-threonine phosphatase interacting protein 1
|
|
chr16_+_75222609
Show fit
|
0.14 |
ENST00000495583.1
|
CTRB1
|
chymotrypsinogen B1
|
|
chr8_+_22054817
Show fit
|
0.14 |
ENST00000432128.5
ENST00000443491.6
ENST00000517600.5
ENST00000523782.6
|
DMTN
|
dematin actin binding protein
|
|
chr17_+_7439504
Show fit
|
0.14 |
ENST00000575331.1
ENST00000293829.9
|
ENSG00000272884.1
FGF11
|
novel transcript
fibroblast growth factor 11
|
|
chr14_+_23372809
Show fit
|
0.13 |
ENST00000397242.2
ENST00000329715.2
|
IL25
|
interleukin 25
|
|
chr6_+_89433164
Show fit
|
0.13 |
ENST00000369408.9
ENST00000447838.6
|
ANKRD6
|
ankyrin repeat domain 6
|
|
chr8_+_32548590
Show fit
|
0.13 |
ENST00000652588.1
ENST00000521670.5
ENST00000287842.7
|
NRG1
|
neuregulin 1
|
|
chr15_-_52295792
Show fit
|
0.13 |
ENST00000261839.12
|
MYO5C
|
myosin VC
|
|
chr5_+_34915644
Show fit
|
0.13 |
ENST00000336767.6
|
BRIX1
|
biogenesis of ribosomes BRX1
|
|
chr14_-_91867529
Show fit
|
0.13 |
ENST00000435962.7
|
TC2N
|
tandem C2 domains, nuclear
|
|
chr2_+_64454506
Show fit
|
0.13 |
ENST00000409537.2
|
LGALSL
|
galectin like
|
|
chr9_+_38392694
Show fit
|
0.12 |
ENST00000377698.4
ENST00000635162.1
|
ALDH1B1
|
aldehyde dehydrogenase 1 family member B1
|
|
chr6_+_30914205
Show fit
|
0.12 |
ENST00000672801.1
ENST00000321897.9
ENST00000625423.2
ENST00000676266.1
ENST00000428017.5
|
VARS2
|
valyl-tRNA synthetase 2, mitochondrial
|
|
chr18_+_46334197
Show fit
|
0.12 |
ENST00000588679.1
ENST00000543885.2
|
RNF165
|
ring finger protein 165
|
|
chr17_-_43953932
Show fit
|
0.12 |
ENST00000592796.1
|
PYY
|
peptide YY
|
|
chr12_+_112938523
Show fit
|
0.12 |
ENST00000679483.1
ENST00000679493.1
|
OAS3
|
2'-5'-oligoadenylate synthetase 3
|
|
chr2_-_20225433
Show fit
|
0.12 |
ENST00000381150.5
|
SDC1
|
syndecan 1
|
|
chr6_+_89433127
Show fit
|
0.12 |
ENST00000339746.9
|
ANKRD6
|
ankyrin repeat domain 6
|
|
chr7_+_151114597
Show fit
|
0.12 |
ENST00000335367.7
|
AGAP3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3
|
|
chr10_+_132397168
Show fit
|
0.12 |
ENST00000631148.2
ENST00000305233.6
|
PWWP2B
|
PWWP domain containing 2B
|
|
chr1_+_23791138
Show fit
|
0.12 |
ENST00000374514.8
ENST00000420982.5
ENST00000374505.6
|
LYPLA2
|
lysophospholipase 2
|
|
chr22_+_25564818
Show fit
|
0.12 |
ENST00000619906.4
|
GRK3
|
G protein-coupled receptor kinase 3
|
|
chr11_+_66480425
Show fit
|
0.11 |
ENST00000541961.5
ENST00000532019.5
ENST00000526515.5
ENST00000530165.5
ENST00000533725.5
|
DPP3
|
dipeptidyl peptidase 3
|
|
chrX_-_24647091
Show fit
|
0.11 |
ENST00000356768.8
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr1_+_111227699
Show fit
|
0.11 |
ENST00000369748.9
|
CHI3L2
|
chitinase 3 like 2
|
|
chr12_+_112938284
Show fit
|
0.11 |
ENST00000681346.1
|
OAS3
|
2'-5'-oligoadenylate synthetase 3
|
|
chr1_+_111227610
Show fit
|
0.11 |
ENST00000369744.6
|
CHI3L2
|
chitinase 3 like 2
|
|
chr2_-_135985568
Show fit
|
0.11 |
ENST00000264161.9
|
DARS1
|
aspartyl-tRNA synthetase 1
|
|
chr2_+_60881515
Show fit
|
0.11 |
ENST00000295025.12
|
REL
|
REL proto-oncogene, NF-kB subunit
|
|
chr3_-_50292404
Show fit
|
0.11 |
ENST00000417626.8
|
IFRD2
|
interferon related developmental regulator 2
|
|
chr16_-_4416621
Show fit
|
0.10 |
ENST00000570645.5
ENST00000574025.5
ENST00000572898.1
ENST00000537233.6
ENST00000571059.5
|
CORO7
|
coronin 7
|
|
chr16_+_2817180
Show fit
|
0.10 |
ENST00000450020.7
|
PRSS21
|
serine protease 21
|
|
chr5_+_136058849
Show fit
|
0.10 |
ENST00000508076.5
|
TGFBI
|
transforming growth factor beta induced
|
|
chr7_+_29806483
Show fit
|
0.10 |
ENST00000409123.5
ENST00000242140.9
|
WIPF3
|
WAS/WASL interacting protein family member 3
|
|
chr19_-_3786363
Show fit
|
0.10 |
ENST00000310132.11
|
MATK
|
megakaryocyte-associated tyrosine kinase
|
|
chr10_-_124092445
Show fit
|
0.10 |
ENST00000346248.7
|
CHST15
|
carbohydrate sulfotransferase 15
|
|
chr1_-_6485941
Show fit
|
0.10 |
ENST00000676287.1
ENST00000400913.6
|
PLEKHG5
|
pleckstrin homology and RhoGEF domain containing G5
|
|
chr1_+_244461386
Show fit
|
0.10 |
ENST00000366533.8
ENST00000428042.1
|
CATSPERE
|
catsper channel auxiliary subunit epsilon
|
|
chr17_-_27800524
Show fit
|
0.10 |
ENST00000313735.11
|
NOS2
|
nitric oxide synthase 2
|
|
chr19_-_14518383
Show fit
|
0.10 |
ENST00000254322.3
ENST00000595139.2
|
DNAJB1
|
DnaJ heat shock protein family (Hsp40) member B1
|
|
chr2_-_38377256
Show fit
|
0.10 |
ENST00000443098.5
ENST00000449130.5
ENST00000651368.1
ENST00000378954.9
ENST00000419554.6
ENST00000451483.1
ENST00000406122.5
|
ATL2
|
atlastin GTPase 2
|
|
chr8_+_22057857
Show fit
|
0.10 |
ENST00000517305.4
ENST00000265800.9
ENST00000517418.5
|
DMTN
|
dematin actin binding protein
|
|
chr12_+_6946468
Show fit
|
0.10 |
ENST00000543115.5
ENST00000399448.5
|
PTPN6
|
protein tyrosine phosphatase non-receptor type 6
|
|
chr9_-_124500986
Show fit
|
0.10 |
ENST00000373587.3
|
NR5A1
|
nuclear receptor subfamily 5 group A member 1
|
|
chr8_+_22579139
Show fit
|
0.10 |
ENST00000397761.6
|
PDLIM2
|
PDZ and LIM domain 2
|
|
chr19_+_44644025
Show fit
|
0.10 |
ENST00000406449.8
|
PVR
|
PVR cell adhesion molecule
|
|
chr2_-_230068905
Show fit
|
0.10 |
ENST00000457406.5
ENST00000295190.9
|
SLC16A14
|
solute carrier family 16 member 14
|
|
chr2_-_88947820
Show fit
|
0.10 |
ENST00000496168.1
|
IGKV1-5
|
immunoglobulin kappa variable 1-5
|
|
chr22_-_38317380
Show fit
|
0.10 |
ENST00000413574.6
|
CSNK1E
|
casein kinase 1 epsilon
|
|
chr1_+_244461270
Show fit
|
0.09 |
ENST00000366534.9
|
CATSPERE
|
catsper channel auxiliary subunit epsilon
|
|
chr19_+_44643902
Show fit
|
0.09 |
ENST00000403059.8
ENST00000425690.8
|
PVR
|
PVR cell adhesion molecule
|
|
chr22_+_25564628
Show fit
|
0.09 |
ENST00000324198.11
|
GRK3
|
G protein-coupled receptor kinase 3
|
|
chr1_+_244461370
Show fit
|
0.09 |
ENST00000366531.7
|
CATSPERE
|
catsper channel auxiliary subunit epsilon
|
|
chr1_-_205775182
Show fit
|
0.09 |
ENST00000446390.6
|
RAB29
|
RAB29, member RAS oncogene family
|
|
chr19_+_3136026
Show fit
|
0.09 |
ENST00000262958.4
|
GNA15
|
G protein subunit alpha 15
|
|
chrX_-_15854743
Show fit
|
0.09 |
ENST00000450644.2
|
AP1S2
|
adaptor related protein complex 1 subunit sigma 2
|
|
chr13_+_73054969
Show fit
|
0.09 |
ENST00000539231.5
|
KLF5
|
Kruppel like factor 5
|
|
chr3_-_197298558
Show fit
|
0.09 |
ENST00000656944.1
ENST00000346964.6
ENST00000448528.6
ENST00000655488.1
ENST00000357674.9
ENST00000667157.1
ENST00000661336.1
ENST00000654737.1
ENST00000659716.1
ENST00000657381.1
|
DLG1
|
discs large MAGUK scaffold protein 1
|
|
chr3_-_50303565
Show fit
|
0.09 |
ENST00000266031.8
ENST00000395143.6
ENST00000457214.6
ENST00000447605.2
ENST00000395144.7
ENST00000418723.1
|
HYAL1
|
hyaluronidase 1
|
|
chr15_+_85380625
Show fit
|
0.09 |
ENST00000560302.5
|
AKAP13
|
A-kinase anchoring protein 13
|
|
chr15_+_89690804
Show fit
|
0.09 |
ENST00000268130.12
ENST00000560294.5
ENST00000558000.1
|
WDR93
|
WD repeat domain 93
|
|
chr1_+_150364136
Show fit
|
0.09 |
ENST00000369068.5
|
RPRD2
|
regulation of nuclear pre-mRNA domain containing 2
|
|
chr19_-_51723968
Show fit
|
0.09 |
ENST00000222115.5
ENST00000540069.7
|
HAS1
|
hyaluronan synthase 1
|
|
chr5_-_78648718
Show fit
|
0.09 |
ENST00000380345.7
|
LHFPL2
|
LHFPL tetraspan subfamily member 2
|
|
chr20_-_49713842
Show fit
|
0.09 |
ENST00000371711.4
|
B4GALT5
|
beta-1,4-galactosyltransferase 5
|
|
chr19_+_44643798
Show fit
|
0.08 |
ENST00000344956.8
|
PVR
|
PVR cell adhesion molecule
|
|
chr8_+_22579100
Show fit
|
0.08 |
ENST00000452226.5
ENST00000397760.8
|
PDLIM2
|
PDZ and LIM domain 2
|
|
chr19_+_58451607
Show fit
|
0.08 |
ENST00000545523.5
ENST00000599194.5
ENST00000336614.9
ENST00000598244.5
ENST00000599193.5
ENST00000594214.1
|
ZNF324B
|
zinc finger protein 324B
|
|
chr9_-_113376973
Show fit
|
0.08 |
ENST00000374180.4
|
HDHD3
|
haloacid dehalogenase like hydrolase domain containing 3
|
|
chr11_+_706222
Show fit
|
0.08 |
ENST00000318562.13
ENST00000533500.5
|
EPS8L2
|
EPS8 like 2
|
|
chr2_+_60881553
Show fit
|
0.08 |
ENST00000394479.4
|
REL
|
REL proto-oncogene, NF-kB subunit
|
|
chr11_+_706117
Show fit
|
0.08 |
ENST00000533256.5
ENST00000614442.4
|
EPS8L2
|
EPS8 like 2
|
|
chr1_-_205775449
Show fit
|
0.08 |
ENST00000235932.8
ENST00000437324.6
ENST00000414729.1
ENST00000367139.8
|
RAB29
|
RAB29, member RAS oncogene family
|
|
chr1_+_202193791
Show fit
|
0.08 |
ENST00000367278.8
|
LGR6
|
leucine rich repeat containing G protein-coupled receptor 6
|
|
chr19_-_4831689
Show fit
|
0.08 |
ENST00000248244.6
|
TICAM1
|
toll like receptor adaptor molecule 1
|
|
chr16_-_4416564
Show fit
|
0.08 |
ENST00000572467.5
ENST00000251166.9
ENST00000572044.1
ENST00000571052.5
|
CORO7-PAM16
CORO7
|
CORO7-PAM16 readthrough
coronin 7
|
|
chr19_-_14529193
Show fit
|
0.08 |
ENST00000596853.6
ENST00000676515.1
ENST00000678338.1
ENST00000595992.6
ENST00000677848.1
ENST00000677762.1
ENST00000678009.1
ENST00000596075.2
ENST00000601533.6
ENST00000396969.8
ENST00000598692.2
ENST00000678098.1
|
DNAJB1
|
DnaJ heat shock protein family (Hsp40) member B1
|
|
chr14_-_50830641
Show fit
|
0.08 |
ENST00000453196.6
ENST00000496749.1
|
NIN
|
ninein
|
|
chr15_+_91853819
Show fit
|
0.08 |
ENST00000424469.2
|
SLCO3A1
|
solute carrier organic anion transporter family member 3A1
|
|
chr2_-_74441882
Show fit
|
0.08 |
ENST00000272430.10
|
RTKN
|
rhotekin
|
|
chrX_+_131058340
Show fit
|
0.08 |
ENST00000276211.10
ENST00000370922.5
|
ARHGAP36
|
Rho GTPase activating protein 36
|
|
chr21_+_31873010
Show fit
|
0.08 |
ENST00000270112.7
|
HUNK
|
hormonally up-regulated Neu-associated kinase
|
|
chr18_+_46334007
Show fit
|
0.08 |
ENST00000269439.12
ENST00000590330.1
|
RNF165
|
ring finger protein 165
|
|
chr19_-_55370455
Show fit
|
0.08 |
ENST00000264563.7
ENST00000585513.1
ENST00000590625.5
|
IL11
|
interleukin 11
|
|
chr8_-_61646807
Show fit
|
0.07 |
ENST00000522919.5
|
ASPH
|
aspartate beta-hydroxylase
|
|
chr8_+_119067239
Show fit
|
0.07 |
ENST00000332843.3
|
COLEC10
|
collectin subfamily member 10
|
|
chr1_+_19251786
Show fit
|
0.07 |
ENST00000330263.5
|
MRTO4
|
MRT4 homolog, ribosome maturation factor
|
|
chr11_+_67583803
Show fit
|
0.07 |
ENST00000398606.10
|
GSTP1
|
glutathione S-transferase pi 1
|
|
chr12_+_112938422
Show fit
|
0.07 |
ENST00000680044.1
ENST00000680966.1
ENST00000548514.2
ENST00000681497.1
ENST00000551007.1
ENST00000228928.12
ENST00000680438.1
ENST00000681147.1
ENST00000679354.1
ENST00000681085.1
ENST00000680161.1
|
OAS3
|
2'-5'-oligoadenylate synthetase 3
|
|
chr6_+_43517079
Show fit
|
0.07 |
ENST00000372344.6
ENST00000642195.1
ENST00000643799.1
ENST00000304004.7
ENST00000646433.1
ENST00000607635.2
ENST00000643341.1
ENST00000423780.1
|
POLR1C
|
RNA polymerase I and III subunit C
|
|
chr11_+_67583742
Show fit
|
0.07 |
ENST00000398603.6
|
GSTP1
|
glutathione S-transferase pi 1
|
|
chr11_+_66480007
Show fit
|
0.07 |
ENST00000531863.5
ENST00000532677.5
|
DPP3
|
dipeptidyl peptidase 3
|
|
chr19_+_1407731
Show fit
|
0.07 |
ENST00000592453.2
|
DAZAP1
|
DAZ associated protein 1
|
|
chr2_+_26692686
Show fit
|
0.07 |
ENST00000620977.1
ENST00000302909.4
|
KCNK3
|
potassium two pore domain channel subfamily K member 3
|
|
chr8_+_22999535
Show fit
|
0.07 |
ENST00000251822.7
|
RHOBTB2
|
Rho related BTB domain containing 2
|
|
chr17_+_12665882
Show fit
|
0.07 |
ENST00000425538.6
|
MYOCD
|
myocardin
|
|
chr8_+_22578735
Show fit
|
0.07 |
ENST00000339162.11
ENST00000308354.11
|
PDLIM2
|
PDZ and LIM domain 2
|
|
chr22_-_37953541
Show fit
|
0.07 |
ENST00000422191.1
ENST00000249079.6
ENST00000403305.6
ENST00000418863.5
ENST00000619227.4
ENST00000403026.5
|
C22orf23
|
chromosome 22 open reading frame 23
|
|
chr19_+_38899946
Show fit
|
0.07 |
ENST00000572515.5
ENST00000313582.6
ENST00000575359.5
|
NFKBIB
|
NFKB inhibitor beta
|
|
chr17_-_3609411
Show fit
|
0.07 |
ENST00000572705.2
|
TRPV1
|
transient receptor potential cation channel subfamily V member 1
|
|
chr11_+_706196
Show fit
|
0.06 |
ENST00000534755.5
ENST00000650127.1
|
EPS8L2
|
EPS8 like 2
|
|
chr19_-_3786408
Show fit
|
0.06 |
ENST00000395040.6
|
MATK
|
megakaryocyte-associated tyrosine kinase
|
|
chr17_-_7290392
Show fit
|
0.06 |
ENST00000571464.1
|
YBX2
|
Y-box binding protein 2
|
|
chr14_+_32329256
Show fit
|
0.06 |
ENST00000280979.9
|
AKAP6
|
A-kinase anchoring protein 6
|
|
chr1_+_23791203
Show fit
|
0.06 |
ENST00000374503.7
ENST00000374502.7
|
LYPLA2
|
lysophospholipase 2
|
|
chr5_+_137867868
Show fit
|
0.06 |
ENST00000515645.1
|
MYOT
|
myotilin
|
|
chr3_+_23805941
Show fit
|
0.06 |
ENST00000306627.8
ENST00000346855.7
|
UBE2E1
|
ubiquitin conjugating enzyme E2 E1
|
|
chr16_-_10580577
Show fit
|
0.06 |
ENST00000359543.8
|
EMP2
|
epithelial membrane protein 2
|
|
chr16_-_74700786
Show fit
|
0.06 |
ENST00000306247.11
ENST00000575686.1
|
MLKL
|
mixed lineage kinase domain like pseudokinase
|
|
chr2_+_218880844
Show fit
|
0.06 |
ENST00000258411.8
|
WNT10A
|
Wnt family member 10A
|
|
chr18_+_58255433
Show fit
|
0.06 |
ENST00000635997.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase
|
|
chr1_+_165895564
Show fit
|
0.06 |
ENST00000469256.6
|
UCK2
|
uridine-cytidine kinase 2
|
|
chr5_-_95961830
Show fit
|
0.06 |
ENST00000513343.1
ENST00000237853.9
|
ELL2
|
elongation factor for RNA polymerase II 2
|
|
chr12_+_122835426
Show fit
|
0.06 |
ENST00000253083.9
|
HIP1R
|
huntingtin interacting protein 1 related
|
|
chr5_-_98928992
Show fit
|
0.06 |
ENST00000614616.5
|
CHD1
|
chromodomain helicase DNA binding protein 1
|
|
chr19_-_55166632
Show fit
|
0.06 |
ENST00000532817.5
ENST00000527223.6
ENST00000391720.8
|
DNAAF3
|
dynein axonemal assembly factor 3
|
|
chr19_+_49591170
Show fit
|
0.06 |
ENST00000418929.7
|
PRR12
|
proline rich 12
|
|
chr3_+_184561768
Show fit
|
0.06 |
ENST00000330394.3
|
EPHB3
|
EPH receptor B3
|
|
chr3_+_114056728
Show fit
|
0.06 |
ENST00000485050.5
ENST00000281273.8
ENST00000479882.5
ENST00000493014.1
|
QTRT2
|
queuine tRNA-ribosyltransferase accessory subunit 2
|
|
chr4_-_932187
Show fit
|
0.06 |
ENST00000618573.4
|
GAK
|
cyclin G associated kinase
|
|
chr13_-_110561668
Show fit
|
0.06 |
ENST00000267328.5
|
RAB20
|
RAB20, member RAS oncogene family
|
|
chr1_-_112956063
Show fit
|
0.06 |
ENST00000538576.5
ENST00000369626.8
ENST00000458229.6
|
SLC16A1
|
solute carrier family 16 member 1
|
|
chr1_+_89821921
Show fit
|
0.06 |
ENST00000394593.7
|
LRRC8D
|
leucine rich repeat containing 8 VRAC subunit D
|
|
chr20_-_4815100
Show fit
|
0.06 |
ENST00000379376.2
|
RASSF2
|
Ras association domain family member 2
|
|
chrX_-_49184789
Show fit
|
0.06 |
ENST00000453382.5
ENST00000432913.5
|
PRICKLE3
|
prickle planar cell polarity protein 3
|
|
chr7_-_111562455
Show fit
|
0.05 |
ENST00000452895.5
ENST00000405709.7
ENST00000452753.1
ENST00000331762.7
|
IMMP2L
|
inner mitochondrial membrane peptidase subunit 2
|
|
chr2_+_86720282
Show fit
|
0.05 |
ENST00000283632.5
|
RMND5A
|
required for meiotic nuclear division 5 homolog A
|
|
chr10_+_12068945
Show fit
|
0.05 |
ENST00000263035.9
|
DHTKD1
|
dehydrogenase E1 and transketolase domain containing 1
|
|
chr16_-_74700845
Show fit
|
0.05 |
ENST00000308807.12
ENST00000573267.1
|
MLKL
|
mixed lineage kinase domain like pseudokinase
|
|
chr17_-_63446168
Show fit
|
0.05 |
ENST00000584031.5
ENST00000392976.5
|
CYB561
|
cytochrome b561
|
|
chr1_+_165895583
Show fit
|
0.05 |
ENST00000470820.1
|
UCK2
|
uridine-cytidine kinase 2
|
|
chr15_+_91853690
Show fit
|
0.05 |
ENST00000318445.11
|
SLCO3A1
|
solute carrier organic anion transporter family member 3A1
|
|
chr19_-_14517425
Show fit
|
0.05 |
ENST00000676577.1
ENST00000677204.1
ENST00000598235.2
|
DNAJB1
|
DnaJ heat shock protein family (Hsp40) member B1
|
|
chr7_+_142332182
Show fit
|
0.05 |
ENST00000547918.2
|
TRBV7-1
|
T cell receptor beta variable 7-1 (non-functional)
|
|
chr4_-_73998669
Show fit
|
0.05 |
ENST00000296027.5
|
CXCL5
|
C-X-C motif chemokine ligand 5
|
|
chr3_-_108589375
Show fit
|
0.05 |
ENST00000625495.1
ENST00000619684.4
ENST00000295746.13
|
CIP2A
|
cellular inhibitor of PP2A
|
|
chr10_+_110497898
Show fit
|
0.05 |
ENST00000369583.4
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr12_+_57522801
Show fit
|
0.05 |
ENST00000355673.8
ENST00000546632.1
ENST00000549623.1
|
MBD6
|
methyl-CpG binding domain protein 6
|
|
chr11_+_65181687
Show fit
|
0.05 |
ENST00000527739.5
ENST00000526966.5
ENST00000533129.5
ENST00000524773.5
|
CAPN1
|
calpain 1
|
|
chr2_+_201182873
Show fit
|
0.05 |
ENST00000360132.7
|
CASP10
|
caspase 10
|
|
chr8_+_22589240
Show fit
|
0.05 |
ENST00000450780.6
ENST00000430850.6
ENST00000447849.2
ENST00000614502.4
ENST00000443561.3
|
ENSG00000248235.6
PDLIM2
|
novel protein
PDZ and LIM domain 2
|
|
chr18_+_33578213
Show fit
|
0.05 |
ENST00000681521.1
ENST00000269197.12
|
ASXL3
|
ASXL transcriptional regulator 3
|
|
chr6_+_29942523
Show fit
|
0.05 |
ENST00000376809.10
ENST00000376802.2
ENST00000638375.1
|
HLA-A
|
major histocompatibility complex, class I, A
|
|
chr3_-_194351290
Show fit
|
0.05 |
ENST00000429275.1
ENST00000323830.4
|
CPN2
|
carboxypeptidase N subunit 2
|
|
chr1_-_32817311
Show fit
|
0.05 |
ENST00000373477.9
ENST00000675785.1
|
YARS1
|
tyrosyl-tRNA synthetase 1
|
|
chr2_-_218292496
Show fit
|
0.05 |
ENST00000258412.8
ENST00000445635.5
ENST00000413976.1
|
TMBIM1
|
transmembrane BAX inhibitor motif containing 1
|
|
chr11_+_1410443
Show fit
|
0.05 |
ENST00000528710.5
|
BRSK2
|
BR serine/threonine kinase 2
|
|
chr19_-_55166671
Show fit
|
0.05 |
ENST00000455045.5
|
DNAAF3
|
dynein axonemal assembly factor 3
|