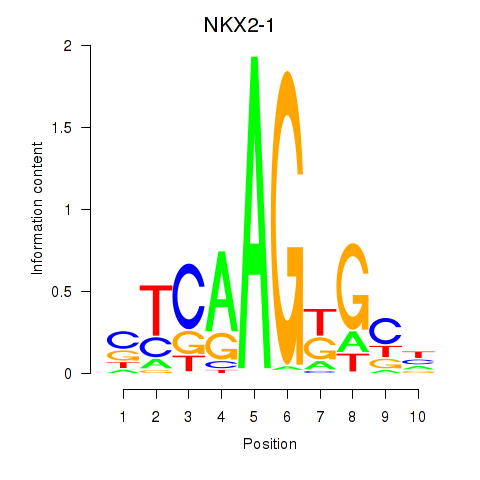
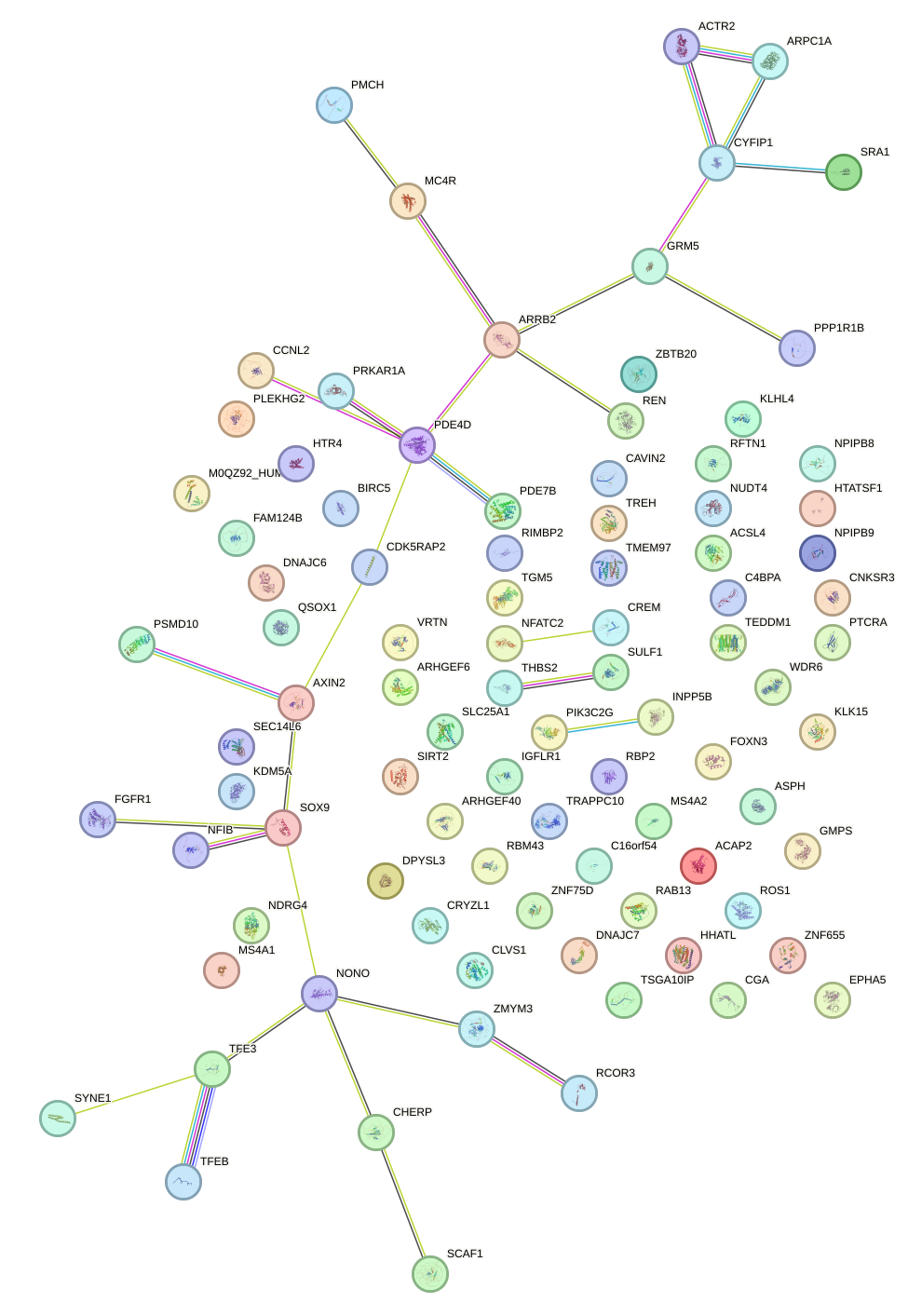
|
chr11_-_118679637
Show fit
|
0.19 |
ENST00000264029.9
ENST00000397925.2
|
TREH
|
trehalase
|
|
chr12_+_18261511
Show fit
|
0.16 |
ENST00000538779.6
ENST00000433979.6
|
PIK3C2G
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma
|
|
chr1_+_180196536
Show fit
|
0.14 |
ENST00000443059.1
|
QSOX1
|
quiescin sulfhydryl oxidase 1
|
|
chr8_-_38468601
Show fit
|
0.13 |
ENST00000341462.9
ENST00000683765.1
ENST00000356207.9
ENST00000326324.10
ENST00000335922.9
ENST00000532791.5
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chrX_+_87517784
Show fit
|
0.12 |
ENST00000373119.9
ENST00000373114.4
|
KLHL4
|
kelch like family member 4
|
|
chr4_-_65670478
Show fit
|
0.11 |
ENST00000613740.5
ENST00000622150.4
ENST00000511294.1
|
EPHA5
|
EPH receptor A5
|
|
chr1_-_153545793
Show fit
|
0.11 |
ENST00000354332.8
ENST00000368716.9
|
S100A4
|
S100 calcium binding protein A4
|
|
chr1_-_153544997
Show fit
|
0.11 |
ENST00000368715.5
|
S100A4
|
S100 calcium binding protein A4
|
|
chr2_-_191847068
Show fit
|
0.10 |
ENST00000304141.5
|
CAVIN2
|
caveolae associated protein 2
|
|
chr6_-_154430495
Show fit
|
0.10 |
ENST00000424998.3
|
CNKSR3
|
CNKSR family member 3
|
|
chr5_-_147510056
Show fit
|
0.10 |
ENST00000343218.10
|
DPYSL3
|
dihydropyrimidinase like 3
|
|
chr12_+_59664677
Show fit
|
0.10 |
ENST00000548610.5
|
SLC16A7
|
solute carrier family 16 member 7
|
|
chrX_-_136767322
Show fit
|
0.09 |
ENST00000370620.5
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor 6
|
|
chr6_-_87095059
Show fit
|
0.09 |
ENST00000369582.6
ENST00000610310.3
ENST00000630630.2
ENST00000627148.3
ENST00000625577.1
|
CGA
|
glycoprotein hormones, alpha polypeptide
|
|
chr6_+_136038195
Show fit
|
0.08 |
ENST00000615259.4
|
PDE7B
|
phosphodiesterase 7B
|
|
chr22_-_19178402
Show fit
|
0.08 |
ENST00000451283.5
|
SLC25A1
|
solute carrier family 25 member 1
|
|
chrX_-_109733249
Show fit
|
0.08 |
ENST00000469796.7
ENST00000672401.1
ENST00000671846.1
|
ACSL4
|
acyl-CoA synthetase long chain family member 4
|
|
chr14_+_21070273
Show fit
|
0.08 |
ENST00000555038.5
ENST00000298694.9
|
ARHGEF40
|
Rho guanine nucleotide exchange factor 40
|
|
chr16_+_58500135
Show fit
|
0.08 |
ENST00000563978.5
ENST00000569923.5
ENST00000356752.8
ENST00000563799.5
ENST00000562999.5
ENST00000570248.5
ENST00000562731.5
ENST00000568424.1
|
NDRG4
|
NDRG family member 4
|
|
chr16_+_58500047
Show fit
|
0.07 |
ENST00000566192.5
ENST00000565088.5
ENST00000568640.5
|
NDRG4
|
NDRG family member 4
|
|
chr1_+_65309517
Show fit
|
0.07 |
ENST00000371069.5
|
DNAJC6
|
DnaJ heat shock protein family (Hsp40) member C6
|
|
chr14_-_89417148
Show fit
|
0.07 |
ENST00000557258.6
|
FOXN3
|
forkhead box N3
|
|
chrX_-_109733220
Show fit
|
0.07 |
ENST00000672282.1
ENST00000340800.7
|
ACSL4
|
acyl-CoA synthetase long chain family member 4
|
|
chr6_-_152637351
Show fit
|
0.07 |
ENST00000367255.10
|
SYNE1
|
spectrin repeat containing nuclear envelope protein 1
|
|
chrX_-_109733181
Show fit
|
0.06 |
ENST00000673016.1
|
ACSL4
|
acyl-CoA synthetase long chain family member 4
|
|
chr19_-_50833187
Show fit
|
0.06 |
ENST00000598673.1
|
KLK15
|
kallikrein related peptidase 15
|
|
chr14_+_74348440
Show fit
|
0.06 |
ENST00000256362.5
|
VRTN
|
vertebrae development associated
|
|
chr3_-_16482850
Show fit
|
0.06 |
ENST00000432519.5
|
RFTN1
|
raftlin, lipid raft linker 1
|
|
chr17_+_39628496
Show fit
|
0.06 |
ENST00000394265.5
ENST00000394267.2
|
PPP1R1B
|
protein phosphatase 1 regulatory inhibitor subunit 1B
|
|
chr19_-_35740535
Show fit
|
0.06 |
ENST00000591748.1
|
IGFLR1
|
IGF like family receptor 1
|
|
chr17_+_4710622
Show fit
|
0.06 |
ENST00000574954.5
ENST00000269260.7
ENST00000346341.6
ENST00000572457.5
ENST00000381488.10
ENST00000412477.7
ENST00000571428.5
ENST00000575877.5
|
ARRB2
|
arrestin beta 2
|
|
chr6_-_41705813
Show fit
|
0.06 |
ENST00000419574.6
ENST00000445214.2
|
TFEB
|
transcription factor EB
|
|
chr2_-_151261839
Show fit
|
0.06 |
ENST00000331426.6
|
RBM43
|
RNA binding motif protein 43
|
|
chrX_+_71254781
Show fit
|
0.06 |
ENST00000677446.1
|
NONO
|
non-POU domain containing octamer binding
|
|
chr7_+_30852273
Show fit
|
0.06 |
ENST00000509504.2
|
ENSG00000250424.4
|
novel protein, MINDY4 and AQP1 readthrough
|
|
chr9_+_35341830
Show fit
|
0.06 |
ENST00000636694.1
|
UNC13B
|
unc-13 homolog B
|
|
chr6_-_117425855
Show fit
|
0.06 |
ENST00000368508.7
|
ROS1
|
ROS proto-oncogene 1, receptor tyrosine kinase
|
|
chr6_-_117425905
Show fit
|
0.06 |
ENST00000368507.8
|
ROS1
|
ROS proto-oncogene 1, receptor tyrosine kinase
|
|
chr5_-_148654522
Show fit
|
0.05 |
ENST00000377888.8
|
HTR4
|
5-hydroxytryptamine receptor 4
|
|
chr1_-_153985366
Show fit
|
0.05 |
ENST00000614713.4
|
RAB13
|
RAB13, member RAS oncogene family
|
|
chr11_+_65945445
Show fit
|
0.05 |
ENST00000532620.5
|
TSGA10IP
|
testis specific 10 interacting protein
|
|
chrX_-_143636094
Show fit
|
0.05 |
ENST00000356928.2
|
SLITRK4
|
SLIT and NTRK like family member 4
|
|
chr9_-_120580125
Show fit
|
0.05 |
ENST00000360190.8
ENST00000349780.9
ENST00000360822.7
|
CDK5RAP2
|
CDK5 regulatory subunit associated protein 2
|
|
chrX_-_135344101
Show fit
|
0.05 |
ENST00000370766.8
|
ZNF75D
|
zinc finger protein 75D
|
|
chr15_-_43266857
Show fit
|
0.05 |
ENST00000349114.8
ENST00000220420.10
|
TGM5
|
transglutaminase 5
|
|
chr8_+_61287950
Show fit
|
0.05 |
ENST00000519846.5
ENST00000325897.5
ENST00000523868.2
ENST00000518592.5
|
CLVS1
|
clavesin 1
|
|
chr2_+_65228122
Show fit
|
0.05 |
ENST00000542850.2
|
ACTR2
|
actin related protein 2
|
|
chr5_-_59039454
Show fit
|
0.05 |
ENST00000358923.10
|
PDE4D
|
phosphodiesterase 4D
|
|
chr5_-_140557414
Show fit
|
0.05 |
ENST00000336283.9
|
SRA1
|
steroid receptor RNA activator 1
|
|
chrX_+_153904714
Show fit
|
0.05 |
ENST00000646375.1
|
AVPR2
|
arginine vasopressin receptor 2
|
|
chr3_-_115071333
Show fit
|
0.05 |
ENST00000462705.5
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chrX_+_136497079
Show fit
|
0.05 |
ENST00000535601.5
ENST00000448450.5
ENST00000425695.5
|
HTATSF1
|
HIV-1 Tat specific factor 1
|
|
chrX_-_71254527
Show fit
|
0.05 |
ENST00000373978.1
ENST00000373981.5
|
ZMYM3
|
zinc finger MYM-type containing 3
|
|
chr1_-_1390943
Show fit
|
0.04 |
ENST00000408952.8
|
CCNL2
|
cyclin L2
|
|
chr3_-_42702778
Show fit
|
0.04 |
ENST00000457462.5
ENST00000441594.6
|
HHATL
|
hedgehog acyltransferase like
|
|
chr3_-_46882106
Show fit
|
0.04 |
ENST00000662933.1
|
MYL3
|
myosin light chain 3
|
|
chr21_+_44012296
Show fit
|
0.04 |
ENST00000291574.9
ENST00000380221.7
|
TRAPPC10
|
trafficking protein particle complex 10
|
|
chr17_-_65561640
Show fit
|
0.04 |
ENST00000618960.4
ENST00000307078.10
|
AXIN2
|
axin 2
|
|
chr1_-_37931712
Show fit
|
0.04 |
ENST00000373027.5
|
INPP5B
|
inositol polyphosphate-5-phosphatase B
|
|
chrX_-_108091520
Show fit
|
0.04 |
ENST00000340200.5
ENST00000217958.8
ENST00000372296.5
ENST00000372295.5
ENST00000361815.9
|
PSMD10
|
proteasome 26S subunit, non-ATPase 10
|
|
chr17_+_78214286
Show fit
|
0.04 |
ENST00000592734.5
ENST00000587746.5
|
BIRC5
|
baculoviral IAP repeat containing 5
|
|
chrX_-_71254106
Show fit
|
0.04 |
ENST00000373984.7
ENST00000314425.9
ENST00000373982.5
|
ZMYM3
|
zinc finger MYM-type containing 3
|
|
chr8_-_61689768
Show fit
|
0.04 |
ENST00000517847.6
ENST00000389204.8
ENST00000517661.5
ENST00000517903.5
ENST00000522603.5
ENST00000541428.5
ENST00000522349.5
ENST00000522835.5
ENST00000518306.5
|
ASPH
|
aspartate beta-hydroxylase
|
|
chr6_-_169250825
Show fit
|
0.04 |
ENST00000676869.1
ENST00000676760.1
|
THBS2
|
thrombospondin 2
|
|
chr19_+_39412650
Show fit
|
0.04 |
ENST00000425673.6
|
PLEKHG2
|
pleckstrin homology and RhoGEF domain containing G2
|
|
chr17_+_78214186
Show fit
|
0.04 |
ENST00000301633.8
ENST00000350051.8
ENST00000374948.6
ENST00000590449.1
|
BIRC5
|
baculoviral IAP repeat containing 5
|
|
chr11_+_6863057
Show fit
|
0.04 |
ENST00000641461.1
|
OR10A2
|
olfactory receptor family 10 subfamily A member 2
|
|
chr9_-_14313642
Show fit
|
0.04 |
ENST00000637742.1
|
NFIB
|
nuclear factor I B
|
|
chr19_+_48469202
Show fit
|
0.04 |
ENST00000427476.4
|
CYTH2
|
cytohesin 2
|
|
chr17_-_42017410
Show fit
|
0.04 |
ENST00000316603.12
ENST00000674175.1
ENST00000674233.1
ENST00000457167.9
ENST00000674306.1
|
DNAJC7
|
DnaJ heat shock protein family (Hsp40) member C7
|
|
chr1_+_211259932
Show fit
|
0.04 |
ENST00000367005.8
|
RCOR3
|
REST corepressor 3
|
|
chr3_+_49007062
Show fit
|
0.03 |
ENST00000395474.7
ENST00000610967.4
ENST00000429900.6
|
WDR6
|
WD repeat domain 6
|
|
chr3_-_195442853
Show fit
|
0.03 |
ENST00000635383.1
ENST00000439666.1
ENST00000618471.4
|
ACAP2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2
|
|
chrX_-_49043345
Show fit
|
0.03 |
ENST00000315869.8
|
TFE3
|
transcription factor binding to IGHM enhancer 3
|
|
chr2_-_224401994
Show fit
|
0.03 |
ENST00000389874.3
|
FAM124B
|
family with sequence similarity 124 member B
|
|
chr3_-_139476508
Show fit
|
0.03 |
ENST00000232217.6
|
RBP2
|
retinol binding protein 2
|
|
chr10_+_35195843
Show fit
|
0.03 |
ENST00000488741.5
ENST00000474931.5
ENST00000468236.5
ENST00000344351.5
ENST00000490511.1
|
CREM
|
cAMP responsive element modulator
|
|
chr21_-_33643926
Show fit
|
0.03 |
ENST00000438788.1
|
CRYZL1
|
crystallin zeta like 1
|
|
chr2_-_224402097
Show fit
|
0.03 |
ENST00000409685.4
|
FAM124B
|
family with sequence similarity 124 member B
|
|
chr22_-_30529163
Show fit
|
0.03 |
ENST00000437871.1
|
SEC14L6
|
SEC14 like lipid binding 6
|
|
chr11_+_60455839
Show fit
|
0.03 |
ENST00000532491.5
ENST00000532073.5
ENST00000345732.9
ENST00000534668.6
ENST00000528313.1
ENST00000533306.6
ENST00000674194.1
|
MS4A1
|
membrane spanning 4-domains A1
|
|
chr7_+_99325857
Show fit
|
0.03 |
ENST00000638617.1
ENST00000262942.10
|
ENSG00000284292.1
ARPC1A
|
novel protein, ARPC1A and ARPC1B readthrough
actin related protein 2/3 complex subunit 1A
|
|
chr3_+_188212931
Show fit
|
0.03 |
ENST00000618621.4
ENST00000640853.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr1_-_182400660
Show fit
|
0.03 |
ENST00000367565.2
|
TEDDM1
|
transmembrane epididymal protein 1
|
|
chr1_+_207104226
Show fit
|
0.03 |
ENST00000367070.8
|
C4BPA
|
complement component 4 binding protein alpha
|
|
chr12_+_93378625
Show fit
|
0.03 |
ENST00000546925.1
|
NUDT4
|
nudix hydrolase 4
|
|
chr17_+_68525795
Show fit
|
0.03 |
ENST00000592800.5
|
PRKAR1A
|
protein kinase cAMP-dependent type I regulatory subunit alpha
|
|
chr11_-_89063631
Show fit
|
0.03 |
ENST00000455756.6
|
GRM5
|
glutamate metabotropic receptor 5
|
|
chr3_+_155870623
Show fit
|
0.03 |
ENST00000295920.7
ENST00000496455.7
|
GMPS
|
guanine monophosphate synthase
|
|
chr7_+_99558632
Show fit
|
0.03 |
ENST00000252713.9
ENST00000449244.5
|
ZNF655
|
zinc finger protein 655
|
|
chr20_-_51562829
Show fit
|
0.03 |
ENST00000609943.5
ENST00000609507.1
|
NFATC2
|
nuclear factor of activated T cells 2
|
|
chr1_-_204166334
Show fit
|
0.03 |
ENST00000272190.9
|
REN
|
renin
|
|
chr16_+_51553436
Show fit
|
0.03 |
ENST00000565308.2
|
HNRNPA1P48
|
heterogeneous nuclear ribonucleoprotein A1 pseudogene 48
|
|
chr19_-_38899529
Show fit
|
0.03 |
ENST00000249396.12
ENST00000437828.5
|
SIRT2
|
sirtuin 2
|
|
chr16_+_28637654
Show fit
|
0.03 |
ENST00000529716.5
|
NPIPB8
|
nuclear pore complex interacting protein family member B8
|
|
chr17_+_28319149
Show fit
|
0.03 |
ENST00000226230.8
ENST00000583381.5
ENST00000582113.1
ENST00000582384.1
|
TMEM97
|
transmembrane protein 97
|
|
chr18_-_60372767
Show fit
|
0.03 |
ENST00000299766.5
|
MC4R
|
melanocortin 4 receptor
|
|
chr16_-_29745951
Show fit
|
0.03 |
ENST00000329410.4
|
C16orf54
|
chromosome 16 open reading frame 54
|
|
chr12_-_102197827
Show fit
|
0.03 |
ENST00000329406.5
|
PMCH
|
pro-melanin concentrating hormone
|
|
chrX_+_153905001
Show fit
|
0.03 |
ENST00000430697.1
ENST00000337474.5
ENST00000370049.1
|
AVPR2
|
arginine vasopressin receptor 2
|
|
chr6_+_42916046
Show fit
|
0.02 |
ENST00000304672.6
|
PTCRA
|
pre T cell antigen receptor alpha
|
|
chr8_+_69492793
Show fit
|
0.02 |
ENST00000616868.1
ENST00000419716.7
ENST00000402687.9
|
SULF1
|
sulfatase 1
|
|
chr1_-_153626948
Show fit
|
0.02 |
ENST00000392622.3
|
S100A13
|
S100 calcium binding protein A13
|
|
chr6_+_42915989
Show fit
|
0.02 |
ENST00000441198.4
ENST00000446507.5
ENST00000616441.2
|
PTCRA
|
pre T cell antigen receptor alpha
|
|
chr17_+_36544896
Show fit
|
0.02 |
ENST00000611219.1
ENST00000613102.5
|
GGNBP2
|
gametogenetin binding protein 2
|
|
chr3_+_39107654
Show fit
|
0.02 |
ENST00000683103.1
ENST00000431162.6
|
TTC21A
|
tetratricopeptide repeat domain 21A
|
|
chr17_-_8295342
Show fit
|
0.02 |
ENST00000579192.5
ENST00000577745.2
|
SLC25A35
|
solute carrier family 25 member 35
|
|
chr7_+_128241272
Show fit
|
0.02 |
ENST00000308868.5
|
LEP
|
leptin
|
|
chr12_-_10826358
Show fit
|
0.02 |
ENST00000240619.2
|
TAS2R10
|
taste 2 receptor member 10
|
|
chr1_-_160262407
Show fit
|
0.02 |
ENST00000419626.1
ENST00000610139.5
ENST00000475733.5
ENST00000407642.6
ENST00000368073.7
ENST00000326837.6
|
DCAF8
|
DDB1 and CUL4 associated factor 8
|
|
chr5_-_135447348
Show fit
|
0.02 |
ENST00000503143.3
|
DCANP1
|
dendritic cell associated nuclear protein
|
|
chr7_-_120857124
Show fit
|
0.02 |
ENST00000441017.5
ENST00000424710.5
ENST00000433758.5
|
TSPAN12
|
tetraspanin 12
|
|
chr17_+_3207539
Show fit
|
0.02 |
ENST00000641322.1
ENST00000641732.2
|
OR1A1
|
olfactory receptor family 1 subfamily A member 1
|
|
chr11_+_106077638
Show fit
|
0.02 |
ENST00000278618.9
|
AASDHPPT
|
aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase
|
|
chr4_+_54657918
Show fit
|
0.02 |
ENST00000412167.6
ENST00000288135.6
|
KIT
|
KIT proto-oncogene, receptor tyrosine kinase
|
|
chr3_-_39107591
Show fit
|
0.02 |
ENST00000319283.8
ENST00000422110.6
ENST00000479927.5
|
GORASP1
|
golgi reassembly stacking protein 1
|
|
chr2_-_240561029
Show fit
|
0.02 |
ENST00000405002.5
ENST00000441168.5
ENST00000403283.5
|
ANKMY1
|
ankyrin repeat and MYND domain containing 1
|
|
chr6_-_49463173
Show fit
|
0.02 |
ENST00000274813.4
|
MMUT
|
methylmalonyl-CoA mutase
|
|
chr4_-_154550376
Show fit
|
0.02 |
ENST00000302078.9
ENST00000499023.7
|
PLRG1
|
pleiotropic regulator 1
|
|
chr17_-_81683782
Show fit
|
0.02 |
ENST00000622299.5
|
ARL16
|
ADP ribosylation factor like GTPase 16
|
|
chr2_-_85668172
Show fit
|
0.02 |
ENST00000428225.5
ENST00000519937.7
|
SFTPB
|
surfactant protein B
|
|
chr1_-_9943314
Show fit
|
0.02 |
ENST00000377223.6
ENST00000377213.1
|
LZIC
|
leucine zipper and CTNNBIP1 domain containing
|
|
chr12_+_93378550
Show fit
|
0.02 |
ENST00000550056.5
ENST00000549992.5
ENST00000548662.5
ENST00000547014.5
|
NUDT4
|
nudix hydrolase 4
|
|
chr14_+_22070548
Show fit
|
0.02 |
ENST00000390450.3
|
TRAV22
|
T cell receptor alpha variable 22
|
|
chr8_-_140800535
Show fit
|
0.02 |
ENST00000521986.5
ENST00000523539.5
|
PTK2
|
protein tyrosine kinase 2
|
|
chr17_-_78782257
Show fit
|
0.02 |
ENST00000591455.5
ENST00000446868.7
ENST00000361101.8
ENST00000589296.5
|
CYTH1
|
cytohesin 1
|
|
chr22_+_24495242
Show fit
|
0.02 |
ENST00000382760.2
ENST00000326010.10
|
UPB1
|
beta-ureidopropionase 1
|
|
chr5_-_173235194
Show fit
|
0.02 |
ENST00000329198.5
ENST00000521848.1
|
NKX2-5
|
NK2 homeobox 5
|
|
chr17_-_81683659
Show fit
|
0.01 |
ENST00000574938.5
ENST00000570561.5
ENST00000573392.5
ENST00000576135.5
ENST00000573715.2
|
ARL16
|
ADP ribosylation factor like GTPase 16
|
|
chrX_+_135344784
Show fit
|
0.01 |
ENST00000370761.7
ENST00000339249.5
|
ZNF449
|
zinc finger protein 449
|
|
chr7_-_27185223
Show fit
|
0.01 |
ENST00000517402.1
ENST00000006015.4
|
HOXA11
|
homeobox A11
|
|
chr16_-_21159441
Show fit
|
0.01 |
ENST00000261383.3
|
DNAH3
|
dynein axonemal heavy chain 3
|
|
chr14_+_80955577
Show fit
|
0.01 |
ENST00000642209.1
ENST00000298171.7
|
TSHR
|
thyroid stimulating hormone receptor
|
|
chr1_+_54998927
Show fit
|
0.01 |
ENST00000651561.1
|
BSND
|
barttin CLCNK type accessory subunit beta
|
|
chr15_+_40161003
Show fit
|
0.01 |
ENST00000412359.7
ENST00000287598.11
|
BUB1B
|
BUB1 mitotic checkpoint serine/threonine kinase B
|
|
chr17_+_42659264
Show fit
|
0.01 |
ENST00000251412.8
|
TUBG2
|
tubulin gamma 2
|
|
chr5_+_140700437
Show fit
|
0.01 |
ENST00000274712.8
|
ZMAT2
|
zinc finger matrin-type 2
|
|
chr19_+_48469354
Show fit
|
0.01 |
ENST00000452733.7
ENST00000641098.1
|
CYTH2
|
cytohesin 2
|
|
chr2_-_156332694
Show fit
|
0.01 |
ENST00000424077.1
ENST00000426264.5
ENST00000421709.1
ENST00000339562.9
|
NR4A2
|
nuclear receptor subfamily 4 group A member 2
|
|
chr17_-_47957824
Show fit
|
0.01 |
ENST00000300557.3
|
PRR15L
|
proline rich 15 like
|
|
chr1_+_150293830
Show fit
|
0.01 |
ENST00000614145.5
ENST00000581066.2
|
MRPS21
|
mitochondrial ribosomal protein S21
|
|
chr5_+_111092329
Show fit
|
0.01 |
ENST00000513710.4
|
WDR36
|
WD repeat domain 36
|
|
chr8_+_144148640
Show fit
|
0.01 |
ENST00000534366.5
|
MROH1
|
maestro heat like repeat family member 1
|
|
chr2_-_175168159
Show fit
|
0.01 |
ENST00000392544.5
ENST00000409499.5
ENST00000409833.5
ENST00000264110.7
ENST00000409635.5
ENST00000345739.9
ENST00000426833.7
|
ATF2
|
activating transcription factor 2
|
|
chr6_-_149484965
Show fit
|
0.01 |
ENST00000409806.8
|
ZC3H12D
|
zinc finger CCCH-type containing 12D
|
|
chr14_+_73886617
Show fit
|
0.01 |
ENST00000540593.5
ENST00000555730.6
|
ZNF410
|
zinc finger protein 410
|
|
chr17_-_8152380
Show fit
|
0.01 |
ENST00000317276.9
|
PER1
|
period circadian regulator 1
|
|
chr3_+_19148500
Show fit
|
0.01 |
ENST00000328405.7
|
KCNH8
|
potassium voltage-gated channel subfamily H member 8
|
|
chr20_+_31819348
Show fit
|
0.01 |
ENST00000375985.5
|
MYLK2
|
myosin light chain kinase 2
|
|
chr2_+_128091166
Show fit
|
0.01 |
ENST00000259253.11
|
UGGT1
|
UDP-glucose glycoprotein glucosyltransferase 1
|
|
chr20_+_31819302
Show fit
|
0.01 |
ENST00000375994.6
|
MYLK2
|
myosin light chain kinase 2
|
|
chr1_-_17011891
Show fit
|
0.01 |
ENST00000341676.9
ENST00000326735.13
ENST00000452699.5
|
ATP13A2
|
ATPase cation transporting 13A2
|
|
chr10_-_79560386
Show fit
|
0.01 |
ENST00000372327.9
ENST00000417041.1
ENST00000640627.1
ENST00000372325.7
|
SFTPA2
|
surfactant protein A2
|
|
chr14_+_80955043
Show fit
|
0.01 |
ENST00000541158.6
|
TSHR
|
thyroid stimulating hormone receptor
|
|
chrX_-_16869840
Show fit
|
0.01 |
ENST00000380084.8
|
RBBP7
|
RB binding protein 7, chromatin remodeling factor
|
|
chr14_-_75660816
Show fit
|
0.01 |
ENST00000256319.7
|
ERG28
|
ergosterol biosynthesis 28 homolog
|
|
chr10_+_62049211
Show fit
|
0.01 |
ENST00000309334.5
|
ARID5B
|
AT-rich interaction domain 5B
|
|
chr10_+_100462969
Show fit
|
0.01 |
ENST00000343737.6
|
WNT8B
|
Wnt family member 8B
|
|
chr12_-_610407
Show fit
|
0.01 |
ENST00000397265.7
|
NINJ2
|
ninjurin 2
|
|
chr1_+_66533575
Show fit
|
0.01 |
ENST00000684751.1
ENST00000683291.1
ENST00000682054.1
ENST00000435165.3
ENST00000684539.1
ENST00000681971.1
ENST00000682476.1
ENST00000684168.1
ENST00000371039.5
|
SGIP1
|
SH3GL interacting endocytic adaptor 1
|
|
chr5_+_34929572
Show fit
|
0.01 |
ENST00000382021.2
|
DNAJC21
|
DnaJ heat shock protein family (Hsp40) member C21
|
|
chr17_-_41467386
Show fit
|
0.01 |
ENST00000225899.4
|
KRT32
|
keratin 32
|
|
chr9_+_129626287
Show fit
|
0.01 |
ENST00000372480.1
|
NTMT1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1
|
|
chr19_+_39971155
Show fit
|
0.01 |
ENST00000157812.7
ENST00000455878.2
|
PSMC4
|
proteasome 26S subunit, ATPase 4
|
|
chr17_+_81683963
Show fit
|
0.01 |
ENST00000676462.1
ENST00000679336.1
ENST00000678196.1
ENST00000677243.1
ENST00000677044.1
ENST00000677109.1
ENST00000677484.1
ENST00000678105.1
ENST00000677209.1
ENST00000329138.9
ENST00000677225.1
ENST00000678866.1
ENST00000676729.1
ENST00000572392.2
ENST00000577012.1
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate
|
|
chr7_-_92134417
Show fit
|
0.01 |
ENST00000003100.13
|
CYP51A1
|
cytochrome P450 family 51 subfamily A member 1
|
|
chr5_+_34929524
Show fit
|
0.01 |
ENST00000648817.1
|
DNAJC21
|
DnaJ heat shock protein family (Hsp40) member C21
|
|
chrX_-_120559889
Show fit
|
0.01 |
ENST00000371323.3
|
CUL4B
|
cullin 4B
|
|
chr2_+_167187364
Show fit
|
0.01 |
ENST00000672671.1
|
XIRP2
|
xin actin binding repeat containing 2
|
|
chr11_-_106077401
Show fit
|
0.01 |
ENST00000526793.5
|
KBTBD3
|
kelch repeat and BTB domain containing 3
|
|
chr9_-_94593810
Show fit
|
0.01 |
ENST00000375337.4
|
FBP2
|
fructose-bisphosphatase 2
|
|
chr18_+_57147065
Show fit
|
0.01 |
ENST00000585477.2
|
BOD1L2
|
biorientation of chromosomes in cell division 1 like 2
|
|
chr2_+_167187283
Show fit
|
0.01 |
ENST00000409605.1
ENST00000409273.6
|
XIRP2
|
xin actin binding repeat containing 2
|
|
chr22_+_35257452
Show fit
|
0.01 |
ENST00000420166.5
ENST00000216106.6
ENST00000455359.5
|
HMGXB4
|
HMG-box containing 4
|
|
chr5_-_172188185
Show fit
|
0.01 |
ENST00000176763.10
|
STK10
|
serine/threonine kinase 10
|
|
chr1_-_41021712
Show fit
|
0.01 |
ENST00000302946.12
ENST00000372613.6
ENST00000439569.2
|
SLFNL1
|
schlafen like 1
|
|
chr10_+_102503956
Show fit
|
0.01 |
ENST00000369902.8
|
SUFU
|
SUFU negative regulator of hedgehog signaling
|
|
chr12_-_21657831
Show fit
|
0.01 |
ENST00000450584.5
ENST00000350669.5
ENST00000673047.2
|
LDHB
|
lactate dehydrogenase B
|
|
chr13_-_44436801
Show fit
|
0.01 |
ENST00000261489.6
|
TSC22D1
|
TSC22 domain family member 1
|
|
chr1_-_41021743
Show fit
|
0.01 |
ENST00000372611.5
|
SLFNL1
|
schlafen like 1
|
|
chr8_+_22161788
Show fit
|
0.01 |
ENST00000521315.5
ENST00000437090.6
ENST00000679463.1
ENST00000520605.5
ENST00000522109.5
ENST00000524255.5
ENST00000523296.1
|
SFTPC
|
surfactant protein C
|
|
chr17_-_49414802
Show fit
|
0.01 |
ENST00000511832.6
ENST00000419140.7
ENST00000617874.5
|
PHB
|
prohibitin
|
|
chr11_+_121129964
Show fit
|
0.01 |
ENST00000645008.1
|
TECTA
|
tectorin alpha
|
|
chr5_+_134758770
Show fit
|
0.01 |
ENST00000628477.2
ENST00000452510.7
ENST00000354283.8
|
DDX46
|
DEAD-box helicase 46
|
|
chr17_-_7204502
Show fit
|
0.01 |
ENST00000486626.8
ENST00000648263.1
|
DLG4
|
discs large MAGUK scaffold protein 4
|
|
chr17_-_81656532
Show fit
|
0.00 |
ENST00000331056.10
|
PDE6G
|
phosphodiesterase 6G
|
|
chr1_+_206470463
Show fit
|
0.00 |
ENST00000581977.7
ENST00000578328.6
ENST00000584998.5
ENST00000579827.6
|
IKBKE
|
inhibitor of nuclear factor kappa B kinase subunit epsilon
|
|
chrX_-_30308333
Show fit
|
0.00 |
ENST00000378963.1
|
NR0B1
|
nuclear receptor subfamily 0 group B member 1
|
|
chr14_+_67241278
Show fit
|
0.00 |
ENST00000676464.1
|
MPP5
|
membrane palmitoylated protein 5
|
|
chr6_-_70303070
Show fit
|
0.00 |
ENST00000370496.3
ENST00000357250.11
|
COL9A1
|
collagen type IX alpha 1 chain
|
|
chr8_+_22161655
Show fit
|
0.00 |
ENST00000318561.7
|
SFTPC
|
surfactant protein C
|
|
chr2_+_135531460
Show fit
|
0.00 |
ENST00000683871.1
ENST00000409478.5
ENST00000264160.8
ENST00000438014.5
|
R3HDM1
|
R3H domain containing 1
|
|
chrX_+_154398367
Show fit
|
0.00 |
ENST00000436473.5
ENST00000344746.8
ENST00000369817.7
ENST00000458500.5
|
RPL10
|
ribosomal protein L10
|
|
chr2_+_219514477
Show fit
|
0.00 |
ENST00000347842.8
ENST00000358078.5
|
ASIC4
|
acid sensing ion channel subunit family member 4
|
|
chr15_+_57219411
Show fit
|
0.00 |
ENST00000543579.5
ENST00000537840.5
ENST00000343827.7
|
TCF12
|
transcription factor 12
|
|
chr19_+_38899680
Show fit
|
0.00 |
ENST00000576510.5
ENST00000392079.7
|
NFKBIB
|
NFKB inhibitor beta
|
|
chr14_+_67241531
Show fit
|
0.00 |
ENST00000678380.1
|
MPP5
|
membrane palmitoylated protein 5
|
|
chr12_-_21657792
Show fit
|
0.00 |
ENST00000396076.5
|
LDHB
|
lactate dehydrogenase B
|
|
chr8_-_93740718
Show fit
|
0.00 |
ENST00000519109.6
|
RBM12B
|
RNA binding motif protein 12B
|
|
chr17_+_55264952
Show fit
|
0.00 |
ENST00000226067.10
|
HLF
|
HLF transcription factor, PAR bZIP family member
|