Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
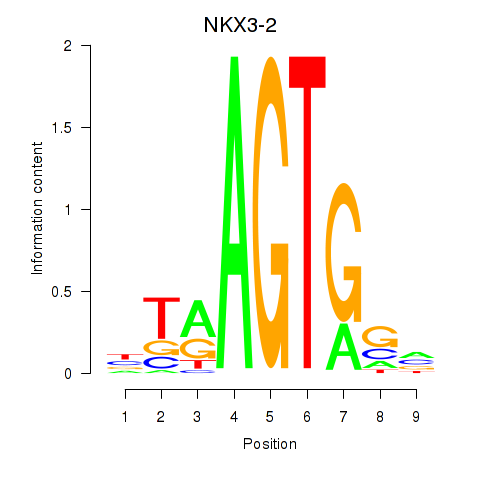
Results for NKX3-2
Z-value: 0.19
Transcription factors associated with NKX3-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX3-2
|
ENSG00000109705.8 | NK3 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX3-2 | hg38_v1_chr4_-_13544506_13544513 | 0.37 | 3.7e-01 | Click! |
Activity profile of NKX3-2 motif
Sorted Z-values of NKX3-2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_96271508 | 0.32 |
ENST00000427367.6
ENST00000413476.6 ENST00000371176.6 |
BLNK
|
B cell linker |
| chr11_-_19202004 | 0.29 |
ENST00000648719.1
|
CSRP3
|
cysteine and glycine rich protein 3 |
| chr12_+_4273751 | 0.24 |
ENST00000675880.1
ENST00000261254.8 |
CCND2
|
cyclin D2 |
| chr10_-_96271553 | 0.22 |
ENST00000224337.10
|
BLNK
|
B cell linker |
| chr11_-_19201976 | 0.21 |
ENST00000647990.1
ENST00000649235.1 ENST00000265968.9 ENST00000649842.1 |
CSRP3
|
cysteine and glycine rich protein 3 |
| chr3_-_112499358 | 0.20 |
ENST00000383680.4
|
BTLA
|
B and T lymphocyte associated |
| chr7_+_150405146 | 0.18 |
ENST00000498682.3
ENST00000641717.1 |
ENSG00000284691.1
|
novel zinc finger protein |
| chr3_-_51974001 | 0.15 |
ENST00000489595.6
ENST00000461108.5 ENST00000395008.6 ENST00000361143.10 ENST00000525795.1 ENST00000488257.2 |
PCBP4
ABHD14B
ENSG00000272762.5
|
poly(rC) binding protein 4 abhydrolase domain containing 14B novel transcript |
| chr3_-_112499457 | 0.15 |
ENST00000334529.10
|
BTLA
|
B and T lymphocyte associated |
| chr6_+_32854179 | 0.14 |
ENST00000374859.3
|
PSMB9
|
proteasome 20S subunit beta 9 |
| chr1_+_151766655 | 0.13 |
ENST00000400999.7
|
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chr14_+_20999255 | 0.13 |
ENST00000554422.5
ENST00000298681.5 |
SLC39A2
|
solute carrier family 39 member 2 |
| chr1_+_205504592 | 0.13 |
ENST00000506784.5
ENST00000360066.6 |
CDK18
|
cyclin dependent kinase 18 |
| chr4_+_113049479 | 0.12 |
ENST00000671727.1
ENST00000671762.1 ENST00000672366.1 ENST00000672502.1 ENST00000672045.1 ENST00000672251.1 ENST00000672854.1 |
ANK2
|
ankyrin 2 |
| chr4_+_113049616 | 0.12 |
ENST00000504454.5
ENST00000357077.9 ENST00000394537.7 ENST00000672779.1 ENST00000264366.10 |
ANK2
|
ankyrin 2 |
| chr9_-_122213874 | 0.11 |
ENST00000482062.1
|
LHX6
|
LIM homeobox 6 |
| chr17_-_7393404 | 0.11 |
ENST00000575434.4
|
PLSCR3
|
phospholipid scramblase 3 |
| chr11_+_59787067 | 0.11 |
ENST00000528805.1
|
STX3
|
syntaxin 3 |
| chr1_+_205504644 | 0.11 |
ENST00000429964.7
ENST00000443813.6 |
CDK18
|
cyclin dependent kinase 18 |
| chr9_-_122213903 | 0.11 |
ENST00000464484.3
|
LHX6
|
LIM homeobox 6 |
| chr1_-_153544997 | 0.10 |
ENST00000368715.5
|
S100A4
|
S100 calcium binding protein A4 |
| chr1_+_14924100 | 0.10 |
ENST00000361144.9
|
KAZN
|
kazrin, periplakin interacting protein |
| chr9_-_92482499 | 0.10 |
ENST00000375544.7
|
ASPN
|
asporin |
| chr12_+_26195647 | 0.10 |
ENST00000535504.1
|
SSPN
|
sarcospan |
| chr2_+_29113989 | 0.10 |
ENST00000404424.5
|
CLIP4
|
CAP-Gly domain containing linker protein family member 4 |
| chr9_-_92482350 | 0.10 |
ENST00000375543.2
|
ASPN
|
asporin |
| chr4_-_158159657 | 0.10 |
ENST00000590648.5
|
GASK1B
|
golgi associated kinase 1B |
| chr1_-_31296748 | 0.10 |
ENST00000263694.9
|
SNRNP40
|
small nuclear ribonucleoprotein U5 subunit 40 |
| chr17_-_81656532 | 0.09 |
ENST00000331056.10
|
PDE6G
|
phosphodiesterase 6G |
| chr11_+_64291285 | 0.09 |
ENST00000422670.7
ENST00000538767.1 |
KCNK4
|
potassium two pore domain channel subfamily K member 4 |
| chr3_+_189631373 | 0.09 |
ENST00000264731.8
ENST00000418709.6 ENST00000320472.9 ENST00000392460.7 ENST00000440651.6 |
TP63
|
tumor protein p63 |
| chr5_+_147878703 | 0.09 |
ENST00000296694.5
|
SCGB3A2
|
secretoglobin family 3A member 2 |
| chr11_-_73142308 | 0.09 |
ENST00000409418.9
|
FCHSD2
|
FCH and double SH3 domains 2 |
| chr1_-_149842736 | 0.09 |
ENST00000369159.2
|
H2AC18
|
H2A clustered histone 18 |
| chr3_-_154430184 | 0.09 |
ENST00000389740.3
|
GPR149
|
G protein-coupled receptor 149 |
| chr1_+_81800906 | 0.09 |
ENST00000674393.1
ENST00000674208.1 |
ADGRL2
|
adhesion G protein-coupled receptor L2 |
| chr14_-_106557465 | 0.08 |
ENST00000390625.3
|
IGHV3-49
|
immunoglobulin heavy variable 3-49 |
| chr17_+_68525795 | 0.08 |
ENST00000592800.5
|
PRKAR1A
|
protein kinase cAMP-dependent type I regulatory subunit alpha |
| chrX_-_31266857 | 0.08 |
ENST00000378702.8
ENST00000361471.8 |
DMD
|
dystrophin |
| chrX_-_15314543 | 0.08 |
ENST00000344384.8
|
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr6_-_53510445 | 0.08 |
ENST00000509541.5
|
GCLC
|
glutamate-cysteine ligase catalytic subunit |
| chr8_-_27258414 | 0.08 |
ENST00000523048.5
|
STMN4
|
stathmin 4 |
| chr1_+_149851053 | 0.08 |
ENST00000607355.2
|
H2AC19
|
H2A clustered histone 19 |
| chr12_-_57767057 | 0.07 |
ENST00000228606.9
|
CYP27B1
|
cytochrome P450 family 27 subfamily B member 1 |
| chr19_-_41436439 | 0.07 |
ENST00000594660.5
|
DMAC2
|
distal membrane arm assembly complex 2 |
| chr5_+_80035341 | 0.07 |
ENST00000350881.6
|
THBS4
|
thrombospondin 4 |
| chr14_-_106639589 | 0.07 |
ENST00000390630.3
|
IGHV4-61
|
immunoglobulin heavy variable 4-61 |
| chr18_-_31760864 | 0.07 |
ENST00000269205.7
ENST00000672005.1 |
SLC25A52
|
solute carrier family 25 member 52 |
| chr10_-_125816596 | 0.07 |
ENST00000368786.5
|
UROS
|
uroporphyrinogen III synthase |
| chr2_+_85584402 | 0.07 |
ENST00000306384.5
|
VAMP5
|
vesicle associated membrane protein 5 |
| chr21_+_29299368 | 0.07 |
ENST00000399921.5
|
BACH1
|
BTB domain and CNC homolog 1 |
| chr10_-_5977492 | 0.07 |
ENST00000530685.5
ENST00000397255.7 ENST00000379971.5 ENST00000528354.5 ENST00000397250.6 ENST00000429135.2 |
IL15RA
|
interleukin 15 receptor subunit alpha |
| chr16_-_138512 | 0.07 |
ENST00000399953.7
|
NPRL3
|
NPR3 like, GATOR1 complex subunit |
| chr2_-_27071628 | 0.07 |
ENST00000447619.5
ENST00000429985.1 ENST00000456793.2 |
OST4
|
oligosaccharyltransferase complex subunit 4, non-catalytic |
| chr17_+_7013638 | 0.07 |
ENST00000547302.3
|
RNASEK-C17orf49
|
RNASEK-C17orf49 readthrough |
| chr8_-_115492221 | 0.07 |
ENST00000518018.1
|
TRPS1
|
transcriptional repressor GATA binding 1 |
| chr4_-_48016631 | 0.07 |
ENST00000513178.2
ENST00000514170.7 |
CNGA1
|
cyclic nucleotide gated channel subunit alpha 1 |
| chr16_+_21678514 | 0.07 |
ENST00000286149.8
ENST00000388958.8 |
OTOA
|
otoancorin |
| chr7_-_22822779 | 0.07 |
ENST00000372879.8
|
TOMM7
|
translocase of outer mitochondrial membrane 7 |
| chr10_+_5048748 | 0.06 |
ENST00000602997.5
ENST00000439082.7 |
AKR1C3
|
aldo-keto reductase family 1 member C3 |
| chr4_-_88823214 | 0.06 |
ENST00000513837.5
ENST00000503556.5 |
FAM13A
|
family with sequence similarity 13 member A |
| chr9_+_112486954 | 0.06 |
ENST00000536272.5
|
KIAA1958
|
KIAA1958 |
| chr8_-_94262308 | 0.06 |
ENST00000297596.3
ENST00000396194.6 |
GEM
|
GTP binding protein overexpressed in skeletal muscle |
| chr19_+_17226662 | 0.06 |
ENST00000598068.5
|
OCEL1
|
occludin/ELL domain containing 1 |
| chr1_-_160282458 | 0.06 |
ENST00000485079.1
|
ENSG00000258465.8
|
novel protein |
| chr16_+_57245336 | 0.06 |
ENST00000562023.5
ENST00000563234.1 |
ARL2BP
|
ADP ribosylation factor like GTPase 2 binding protein |
| chr11_-_107018431 | 0.06 |
ENST00000282249.6
|
GUCY1A2
|
guanylate cyclase 1 soluble subunit alpha 2 |
| chr6_-_136466858 | 0.06 |
ENST00000544465.5
|
MAP7
|
microtubule associated protein 7 |
| chr11_-_120138104 | 0.06 |
ENST00000341846.10
|
TRIM29
|
tripartite motif containing 29 |
| chr22_-_29555178 | 0.06 |
ENST00000418021.1
|
THOC5
|
THO complex 5 |
| chrX_+_22136552 | 0.06 |
ENST00000682888.1
ENST00000684356.1 |
PHEX
|
phosphate regulating endopeptidase homolog X-linked |
| chr19_-_49061997 | 0.06 |
ENST00000593537.1
|
NTF4
|
neurotrophin 4 |
| chr19_+_54105923 | 0.06 |
ENST00000420296.1
|
NDUFA3
|
NADH:ubiquinone oxidoreductase subunit A3 |
| chr3_+_111978996 | 0.06 |
ENST00000273359.8
ENST00000494817.1 |
ABHD10
|
abhydrolase domain containing 10, depalmitoylase |
| chr6_-_32589833 | 0.06 |
ENST00000360004.5
|
HLA-DRB1
|
major histocompatibility complex, class II, DR beta 1 |
| chr7_-_102478895 | 0.06 |
ENST00000393794.4
ENST00000292614.9 |
POLR2J
|
RNA polymerase II subunit J |
| chr19_+_48364361 | 0.05 |
ENST00000344846.7
|
SYNGR4
|
synaptogyrin 4 |
| chr15_-_82806054 | 0.05 |
ENST00000541889.1
ENST00000334574.12 ENST00000561368.1 |
FSD2
|
fibronectin type III and SPRY domain containing 2 |
| chr18_+_26226417 | 0.05 |
ENST00000269142.10
|
TAF4B
|
TATA-box binding protein associated factor 4b |
| chr11_+_44726811 | 0.05 |
ENST00000533202.5
ENST00000520358.7 ENST00000533080.5 ENST00000520999.6 |
TSPAN18
|
tetraspanin 18 |
| chr6_+_42960742 | 0.05 |
ENST00000372808.4
|
GNMT
|
glycine N-methyltransferase |
| chr17_+_60677822 | 0.05 |
ENST00000407086.8
ENST00000589222.5 ENST00000626960.2 ENST00000390652.9 |
BCAS3
|
BCAS3 microtubule associated cell migration factor |
| chr7_-_102517755 | 0.05 |
ENST00000306682.6
ENST00000465829.6 ENST00000541662.5 |
RASA4B
|
RAS p21 protein activator 4B |
| chrX_+_136305988 | 0.05 |
ENST00000394141.1
|
ADGRG4
|
adhesion G protein-coupled receptor G4 |
| chr15_-_93073111 | 0.05 |
ENST00000557420.1
ENST00000542321.6 |
RGMA
|
repulsive guidance molecule BMP co-receptor a |
| chr20_+_2295994 | 0.05 |
ENST00000381458.6
|
TGM3
|
transglutaminase 3 |
| chr9_-_129178247 | 0.05 |
ENST00000372491.4
|
IER5L
|
immediate early response 5 like |
| chr16_+_2205446 | 0.05 |
ENST00000562352.5
ENST00000569417.6 ENST00000562479.5 ENST00000301724.14 |
MLST8
|
MTOR associated protein, LST8 homolog |
| chr7_-_6348906 | 0.05 |
ENST00000313324.9
ENST00000530143.1 |
FAM220A
|
family with sequence similarity 220 member A |
| chr11_-_120123026 | 0.05 |
ENST00000533302.5
|
TRIM29
|
tripartite motif containing 29 |
| chr14_-_34462223 | 0.05 |
ENST00000298130.5
|
SPTSSA
|
serine palmitoyltransferase small subunit A |
| chr20_+_15196834 | 0.05 |
ENST00000402914.5
|
MACROD2
|
mono-ADP ribosylhydrolase 2 |
| chr19_-_6459735 | 0.05 |
ENST00000334510.9
ENST00000301454.9 |
SLC25A23
|
solute carrier family 25 member 23 |
| chr16_+_57372465 | 0.05 |
ENST00000563383.1
|
CX3CL1
|
C-X3-C motif chemokine ligand 1 |
| chr16_+_57372481 | 0.05 |
ENST00000006053.7
|
CX3CL1
|
C-X3-C motif chemokine ligand 1 |
| chr6_-_39322540 | 0.05 |
ENST00000425054.6
ENST00000373227.8 ENST00000373229.9 |
KCNK16
|
potassium two pore domain channel subfamily K member 16 |
| chr3_-_146544636 | 0.05 |
ENST00000486631.5
|
PLSCR1
|
phospholipid scramblase 1 |
| chr11_-_102530738 | 0.05 |
ENST00000260227.5
|
MMP7
|
matrix metallopeptidase 7 |
| chr10_+_129467178 | 0.05 |
ENST00000306010.8
ENST00000651593.1 |
MGMT
|
O-6-methylguanine-DNA methyltransferase |
| chr9_-_23821809 | 0.05 |
ENST00000544538.5
|
ELAVL2
|
ELAV like RNA binding protein 2 |
| chr1_+_86424154 | 0.05 |
ENST00000370565.5
|
CLCA2
|
chloride channel accessory 2 |
| chr1_-_173917281 | 0.05 |
ENST00000367698.4
|
SERPINC1
|
serpin family C member 1 |
| chrX_-_31178220 | 0.05 |
ENST00000681026.1
|
DMD
|
dystrophin |
| chr9_+_33817126 | 0.05 |
ENST00000263228.4
|
UBE2R2
|
ubiquitin conjugating enzyme E2 R2 |
| chr11_+_60455839 | 0.05 |
ENST00000532491.5
ENST00000532073.5 ENST00000345732.9 ENST00000534668.6 ENST00000528313.1 ENST00000533306.6 ENST00000674194.1 |
MS4A1
|
membrane spanning 4-domains A1 |
| chr7_+_130266847 | 0.05 |
ENST00000222481.9
|
CPA2
|
carboxypeptidase A2 |
| chr4_+_143381939 | 0.05 |
ENST00000505913.5
|
GAB1
|
GRB2 associated binding protein 1 |
| chrX_-_31266925 | 0.05 |
ENST00000680557.1
ENST00000378680.6 ENST00000378723.7 ENST00000680768.2 ENST00000681870.1 ENST00000680355.1 ENST00000682322.1 ENST00000682600.1 ENST00000680162.2 ENST00000681153.1 ENST00000681334.1 |
DMD
|
dystrophin |
| chrX_-_31178149 | 0.05 |
ENST00000679437.1
|
DMD
|
dystrophin |
| chr17_+_18476737 | 0.05 |
ENST00000581545.5
ENST00000582333.5 ENST00000328114.11 ENST00000583322.5 ENST00000584941.5 |
LGALS9C
|
galectin 9C |
| chr4_-_167234266 | 0.05 |
ENST00000511269.5
ENST00000506697.5 ENST00000512042.1 |
SPOCK3
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 3 |
| chr2_-_24793382 | 0.05 |
ENST00000328379.6
|
PTRHD1
|
peptidyl-tRNA hydrolase domain containing 1 |
| chr16_+_67279508 | 0.05 |
ENST00000379344.8
ENST00000568621.1 ENST00000450733.5 ENST00000567938.1 |
PLEKHG4
|
pleckstrin homology and RhoGEF domain containing G4 |
| chr1_+_35883189 | 0.04 |
ENST00000674304.1
ENST00000373204.6 ENST00000674426.1 |
AGO1
|
argonaute RISC component 1 |
| chr10_-_27240505 | 0.04 |
ENST00000375888.5
ENST00000676732.1 |
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr22_-_17773976 | 0.04 |
ENST00000317361.11
|
BID
|
BH3 interacting domain death agonist |
| chr1_+_156786875 | 0.04 |
ENST00000526188.5
ENST00000454659.1 |
PRCC
|
proline rich mitotic checkpoint control factor |
| chr6_-_31730198 | 0.04 |
ENST00000375787.6
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr15_-_63157991 | 0.04 |
ENST00000411926.1
|
RPS27L
|
ribosomal protein S27 like |
| chr16_-_2268054 | 0.04 |
ENST00000561518.5
ENST00000561718.5 ENST00000567147.5 ENST00000562690.5 ENST00000569598.6 |
RNPS1
|
RNA binding protein with serine rich domain 1 |
| chr19_-_52171443 | 0.04 |
ENST00000598982.5
ENST00000594362.1 ENST00000597252.5 |
ENSG00000267827.5
ZNF836
|
novel transcript zinc finger protein 836 |
| chr1_-_150876697 | 0.04 |
ENST00000515192.5
|
ARNT
|
aryl hydrocarbon receptor nuclear translocator |
| chr7_-_102616692 | 0.04 |
ENST00000521076.5
ENST00000462172.5 ENST00000522801.5 ENST00000262940.12 ENST00000449970.6 |
RASA4
|
RAS p21 protein activator 4 |
| chr15_-_93073706 | 0.04 |
ENST00000425933.6
|
RGMA
|
repulsive guidance molecule BMP co-receptor a |
| chr19_-_10517439 | 0.04 |
ENST00000333430.6
|
S1PR5
|
sphingosine-1-phosphate receptor 5 |
| chrX_+_48802156 | 0.04 |
ENST00000643374.1
ENST00000644068.1 ENST00000441703.6 ENST00000643934.1 ENST00000489352.5 |
HDAC6
|
histone deacetylase 6 |
| chr1_+_95117923 | 0.04 |
ENST00000455656.1
ENST00000604534.5 |
TLCD4
TLCD4-RWDD3
|
TLC domain containing 4 TLCD4-RWDD3 readthrough |
| chr14_-_55411817 | 0.04 |
ENST00000247178.6
|
ATG14
|
autophagy related 14 |
| chr14_-_106627685 | 0.04 |
ENST00000390629.3
|
IGHV4-59
|
immunoglobulin heavy variable 4-59 |
| chr12_+_123973197 | 0.04 |
ENST00000392404.7
ENST00000337815.9 ENST00000538932.6 ENST00000618862.2 ENST00000389727.8 |
ZNF664
ENSG00000274874.2
RFLNA
|
zinc finger protein 664 novel protein refilin A |
| chr1_-_204494752 | 0.04 |
ENST00000684373.1
|
PIK3C2B
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta |
| chr9_-_72364504 | 0.04 |
ENST00000237937.7
ENST00000343431.6 ENST00000376956.3 |
ZFAND5
|
zinc finger AN1-type containing 5 |
| chr12_-_117361641 | 0.04 |
ENST00000618760.4
|
NOS1
|
nitric oxide synthase 1 |
| chr7_-_727611 | 0.04 |
ENST00000403562.5
|
PRKAR1B
|
protein kinase cAMP-dependent type I regulatory subunit beta |
| chr15_+_81182579 | 0.04 |
ENST00000302987.9
|
IL16
|
interleukin 16 |
| chr2_+_223051814 | 0.04 |
ENST00000281830.3
|
KCNE4
|
potassium voltage-gated channel subfamily E regulatory subunit 4 |
| chr12_+_54008961 | 0.04 |
ENST00000040584.6
|
HOXC8
|
homeobox C8 |
| chr17_-_44389594 | 0.04 |
ENST00000262407.6
|
ITGA2B
|
integrin subunit alpha 2b |
| chr20_+_44355692 | 0.04 |
ENST00000316673.8
ENST00000609795.5 ENST00000457232.5 ENST00000609262.5 |
HNF4A
|
hepatocyte nuclear factor 4 alpha |
| chr1_-_243843164 | 0.04 |
ENST00000491219.6
ENST00000680056.1 ENST00000492957.2 |
AKT3
|
AKT serine/threonine kinase 3 |
| chr15_-_89221558 | 0.04 |
ENST00000268125.10
|
RLBP1
|
retinaldehyde binding protein 1 |
| chr1_-_46616804 | 0.04 |
ENST00000531769.6
ENST00000319928.8 |
MKNK1
MOB3C
|
MAPK interacting serine/threonine kinase 1 MOB kinase activator 3C |
| chr19_+_38619082 | 0.04 |
ENST00000614624.4
ENST00000593149.5 ENST00000538434.5 ENST00000588934.5 ENST00000248342.9 ENST00000545173.6 ENST00000591409.5 ENST00000592558.1 |
EIF3K
|
eukaryotic translation initiation factor 3 subunit K |
| chr16_-_2268111 | 0.04 |
ENST00000301730.12
ENST00000564822.1 ENST00000320225.10 |
RNPS1
|
RNA binding protein with serine rich domain 1 |
| chr8_+_22570944 | 0.04 |
ENST00000517962.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr12_-_122227491 | 0.04 |
ENST00000475784.1
ENST00000645606.1 |
ENSG00000284934.2
|
novel protein |
| chr10_+_5094405 | 0.04 |
ENST00000380554.5
|
AKR1C3
|
aldo-keto reductase family 1 member C3 |
| chr4_+_61202142 | 0.04 |
ENST00000514591.5
|
ADGRL3
|
adhesion G protein-coupled receptor L3 |
| chr15_-_52569197 | 0.04 |
ENST00000563277.5
ENST00000566423.5 |
ARPP19
|
cAMP regulated phosphoprotein 19 |
| chr1_-_24930263 | 0.04 |
ENST00000308873.11
|
RUNX3
|
RUNX family transcription factor 3 |
| chr10_+_103245887 | 0.03 |
ENST00000441178.2
|
RPEL1
|
ribulose-5-phosphate-3-epimerase like 1 |
| chr11_+_93741591 | 0.03 |
ENST00000528288.5
ENST00000617482.4 ENST00000540113.5 |
C11orf54
|
chromosome 11 open reading frame 54 |
| chr5_+_150671588 | 0.03 |
ENST00000523553.1
|
MYOZ3
|
myozenin 3 |
| chr19_-_10333512 | 0.03 |
ENST00000617231.5
ENST00000611074.4 ENST00000615032.4 |
RAVER1
|
ribonucleoprotein, PTB binding 1 |
| chr1_+_164559173 | 0.03 |
ENST00000420696.7
|
PBX1
|
PBX homeobox 1 |
| chr12_-_117361614 | 0.03 |
ENST00000317775.11
|
NOS1
|
nitric oxide synthase 1 |
| chr3_+_186996444 | 0.03 |
ENST00000676633.1
|
ST6GAL1
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 1 |
| chr10_-_27240743 | 0.03 |
ENST00000677901.1
ENST00000677960.1 ENST00000677440.1 ENST00000396271.8 ENST00000677141.1 ENST00000677311.1 ENST00000677667.1 ENST00000677200.1 ENST00000676997.1 ENST00000676511.1 |
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr1_+_119414931 | 0.03 |
ENST00000543831.5
ENST00000433745.5 ENST00000369416.4 |
HSD3B2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr2_+_135531460 | 0.03 |
ENST00000683871.1
ENST00000409478.5 ENST00000264160.8 ENST00000438014.5 |
R3HDM1
|
R3H domain containing 1 |
| chr2_-_207624983 | 0.03 |
ENST00000448007.6
ENST00000432416.5 ENST00000411432.5 ENST00000458426.5 ENST00000406927.6 ENST00000425132.5 |
METTL21A
|
methyltransferase like 21A |
| chr3_-_42875871 | 0.03 |
ENST00000316161.6
ENST00000437102.1 |
CYP8B1
|
cytochrome P450 family 8 subfamily B member 1 |
| chr1_+_151060357 | 0.03 |
ENST00000368921.5
|
MLLT11
|
MLLT11 transcription factor 7 cofactor |
| chr3_-_146544701 | 0.03 |
ENST00000487389.5
|
PLSCR1
|
phospholipid scramblase 1 |
| chr14_+_39114289 | 0.03 |
ENST00000396249.7
ENST00000250379.13 ENST00000534684.7 ENST00000308317.12 ENST00000527381.2 |
GEMIN2
|
gem nuclear organelle associated protein 2 |
| chr4_-_167234426 | 0.03 |
ENST00000541354.5
ENST00000509854.5 ENST00000512681.5 ENST00000357545.9 ENST00000510741.5 ENST00000510403.5 |
SPOCK3
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 3 |
| chr12_+_57941499 | 0.03 |
ENST00000300145.4
|
ATP23
|
ATP23 metallopeptidase and ATP synthase assembly factor homolog |
| chr2_-_70553638 | 0.03 |
ENST00000444975.5
ENST00000445399.5 ENST00000295400.11 ENST00000418333.6 |
TGFA
|
transforming growth factor alpha |
| chr19_+_15086980 | 0.03 |
ENST00000209540.2
|
OR1I1
|
olfactory receptor family 1 subfamily I member 1 |
| chr18_+_58149314 | 0.03 |
ENST00000435432.6
ENST00000357895.9 ENST00000586263.5 |
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr6_-_31729478 | 0.03 |
ENST00000436437.2
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr17_+_55751378 | 0.03 |
ENST00000325214.10
|
PCTP
|
phosphatidylcholine transfer protein |
| chr19_+_18433234 | 0.03 |
ENST00000599699.2
|
SSBP4
|
single stranded DNA binding protein 4 |
| chr6_-_11807045 | 0.03 |
ENST00000379415.6
|
ADTRP
|
androgen dependent TFPI regulating protein |
| chr4_+_155903688 | 0.03 |
ENST00000536354.3
|
TDO2
|
tryptophan 2,3-dioxygenase |
| chr19_-_5680191 | 0.03 |
ENST00000590389.5
|
MICOS13
|
mitochondrial contact site and cristae organizing system subunit 13 |
| chr6_-_31729785 | 0.03 |
ENST00000416410.6
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr2_+_231707650 | 0.03 |
ENST00000409321.5
|
PTMA
|
prothymosin alpha |
| chrX_+_10015226 | 0.03 |
ENST00000380861.9
|
WWC3
|
WWC family member 3 |
| chr1_-_225567270 | 0.03 |
ENST00000497899.6
|
ENAH
|
ENAH actin regulator |
| chr6_-_116060859 | 0.03 |
ENST00000606080.2
|
FRK
|
fyn related Src family tyrosine kinase |
| chr10_+_100535927 | 0.03 |
ENST00000299163.7
|
HIF1AN
|
hypoxia inducible factor 1 subunit alpha inhibitor |
| chr15_-_51243011 | 0.03 |
ENST00000405913.7
ENST00000559878.5 |
CYP19A1
|
cytochrome P450 family 19 subfamily A member 1 |
| chr16_-_70695593 | 0.03 |
ENST00000567648.1
|
VAC14
|
VAC14 component of PIKFYVE complex |
| chr9_-_120793343 | 0.03 |
ENST00000684047.1
|
FBXW2
|
F-box and WD repeat domain containing 2 |
| chr1_-_153985366 | 0.03 |
ENST00000614713.4
|
RAB13
|
RAB13, member RAS oncogene family |
| chr6_+_159790448 | 0.03 |
ENST00000367034.5
|
MRPL18
|
mitochondrial ribosomal protein L18 |
| chrX_+_103628692 | 0.03 |
ENST00000372626.7
|
TCEAL1
|
transcription elongation factor A like 1 |
| chr17_-_42028662 | 0.03 |
ENST00000461831.1
|
ZNF385C
|
zinc finger protein 385C |
| chr1_-_151190125 | 0.03 |
ENST00000354473.4
ENST00000368892.9 ENST00000640458.1 |
VPS72
|
vacuolar protein sorting 72 homolog |
| chr11_+_925840 | 0.03 |
ENST00000448903.7
ENST00000525796.5 ENST00000534328.5 |
AP2A2
|
adaptor related protein complex 2 subunit alpha 2 |
| chr9_-_120793377 | 0.03 |
ENST00000684001.1
ENST00000684405.1 ENST00000608872.6 |
FBXW2
|
F-box and WD repeat domain containing 2 |
| chr10_+_23095556 | 0.03 |
ENST00000376510.8
|
MSRB2
|
methionine sulfoxide reductase B2 |
| chrX_+_153411472 | 0.03 |
ENST00000338647.7
|
ZFP92
|
ZFP92 zinc finger protein |
| chr8_+_21966215 | 0.03 |
ENST00000433566.8
|
XPO7
|
exportin 7 |
| chr12_-_95003666 | 0.03 |
ENST00000327772.7
ENST00000547157.1 ENST00000684171.1 ENST00000547986.5 |
NDUFA12
|
NADH:ubiquinone oxidoreductase subunit A12 |
| chr15_-_56918571 | 0.03 |
ENST00000559000.6
|
ENSG00000285253.1
|
novel protein |
| chr7_-_143647646 | 0.03 |
ENST00000636941.1
|
TCAF2C
|
TRPM8 channel associated factor 2C |
| chr5_+_40909490 | 0.03 |
ENST00000313164.10
|
C7
|
complement C7 |
| chr1_+_158289916 | 0.03 |
ENST00000368170.8
|
CD1C
|
CD1c molecule |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX3-2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.1 | GO:0015847 | putrescine transport(GO:0015847) |
| 0.0 | 0.1 | GO:0016107 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.0 | 0.2 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.1 | GO:0051040 | regulation of calcium-independent cell-cell adhesion(GO:0051040) |
| 0.0 | 0.1 | GO:0071469 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) cellular response to alkaline pH(GO:0071469) |
| 0.0 | 0.1 | GO:0001546 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.0 | 0.1 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.1 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.0 | 0.2 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.1 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.0 | 0.1 | GO:0048867 | stem cell fate determination(GO:0048867) |
| 0.0 | 0.1 | GO:0070843 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 | 0.1 | GO:0099543 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 0.0 | 0.1 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.0 | 0.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:0097274 | urea homeostasis(GO:0097274) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.2 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.1 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.0 | 0.1 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.0 | 0.0 | GO:0061182 | negative regulation of chondrocyte development(GO:0061182) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.1 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.0 | 0.1 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.2 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.1 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.0 | 0.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0015203 | polyamine transmembrane transporter activity(GO:0015203) putrescine transmembrane transporter activity(GO:0015489) |
| 0.0 | 0.5 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.1 | GO:0045550 | geranylgeranyl reductase activity(GO:0045550) delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.0 | 0.1 | GO:0097604 | temperature-gated cation channel activity(GO:0097604) |
| 0.0 | 0.5 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.0 | 0.1 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 0.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 0.1 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.0 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.1 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.0 | GO:0004750 | ribulose-phosphate 3-epimerase activity(GO:0004750) |
| 0.0 | 0.1 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |