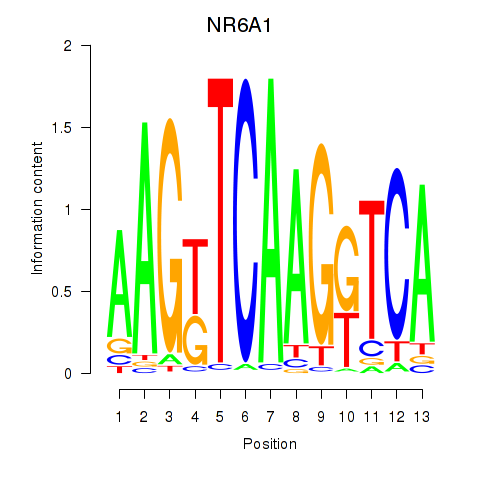
|
chr3_-_151316795
Show fit
|
0.23 |
ENST00000260843.5
|
GPR87
|
G protein-coupled receptor 87
|
|
chr19_-_6720641
Show fit
|
0.23 |
ENST00000245907.11
|
C3
|
complement C3
|
|
chr20_+_59577463
Show fit
|
0.22 |
ENST00000359926.7
|
PHACTR3
|
phosphatase and actin regulator 3
|
|
chr1_-_184974477
Show fit
|
0.19 |
ENST00000367511.4
|
NIBAN1
|
niban apoptosis regulator 1
|
|
chr11_-_108593738
Show fit
|
0.18 |
ENST00000525344.5
ENST00000265843.9
|
EXPH5
|
exophilin 5
|
|
chr10_-_14572123
Show fit
|
0.18 |
ENST00000378465.7
ENST00000452706.6
ENST00000622567.4
ENST00000378458.6
|
FAM107B
|
family with sequence similarity 107 member B
|
|
chr1_-_153544997
Show fit
|
0.18 |
ENST00000368715.5
|
S100A4
|
S100 calcium binding protein A4
|
|
chr5_+_55851378
Show fit
|
0.17 |
ENST00000396836.6
ENST00000359040.10
|
IL31RA
|
interleukin 31 receptor A
|
|
chr17_-_31297231
Show fit
|
0.17 |
ENST00000247271.5
|
OMG
|
oligodendrocyte myelin glycoprotein
|
|
chr11_-_18248662
Show fit
|
0.16 |
ENST00000256733.9
|
SAA2
|
serum amyloid A2
|
|
chr11_-_16408853
Show fit
|
0.16 |
ENST00000528252.5
|
SOX6
|
SRY-box transcription factor 6
|
|
chr5_+_55851349
Show fit
|
0.16 |
ENST00000652347.2
|
IL31RA
|
interleukin 31 receptor A
|
|
chr19_-_15934521
Show fit
|
0.15 |
ENST00000402119.9
|
CYP4F11
|
cytochrome P450 family 4 subfamily F member 11
|
|
chr15_+_100879822
Show fit
|
0.14 |
ENST00000329841.10
ENST00000557963.1
ENST00000346623.6
|
ALDH1A3
|
aldehyde dehydrogenase 1 family member A3
|
|
chr1_+_107139996
Show fit
|
0.14 |
ENST00000370073.6
ENST00000370074.8
|
NTNG1
|
netrin G1
|
|
chr22_+_39994926
Show fit
|
0.14 |
ENST00000333407.11
|
FAM83F
|
family with sequence similarity 83 member F
|
|
chr20_+_45174894
Show fit
|
0.14 |
ENST00000243924.4
|
PI3
|
peptidase inhibitor 3
|
|
chr1_+_151766655
Show fit
|
0.13 |
ENST00000400999.7
|
OAZ3
|
ornithine decarboxylase antizyme 3
|
|
chr8_-_92095627
Show fit
|
0.13 |
ENST00000517919.5
ENST00000617740.4
ENST00000613302.4
ENST00000436581.6
ENST00000614812.4
ENST00000519847.5
|
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1
|
|
chr6_+_30888672
Show fit
|
0.13 |
ENST00000446312.5
|
DDR1
|
discoidin domain receptor tyrosine kinase 1
|
|
chr10_+_24449426
Show fit
|
0.13 |
ENST00000307544.10
|
KIAA1217
|
KIAA1217
|
|
chr11_-_18248632
Show fit
|
0.12 |
ENST00000524555.3
ENST00000528349.5
ENST00000526900.1
ENST00000529528.5
ENST00000414546.6
|
SAA2-SAA4
SAA2
|
SAA2-SAA4 readthrough
serum amyloid A2
|
|
chr12_+_10213417
Show fit
|
0.12 |
ENST00000546017.5
ENST00000535576.5
ENST00000539170.5
|
GABARAPL1
|
GABA type A receptor associated protein like 1
|
|
chr17_+_50426242
Show fit
|
0.12 |
ENST00000502667.5
|
ACSF2
|
acyl-CoA synthetase family member 2
|
|
chr10_+_84173793
Show fit
|
0.11 |
ENST00000372126.4
|
C10orf99
|
chromosome 10 open reading frame 99
|
|
chr8_-_17697654
Show fit
|
0.11 |
ENST00000297488.10
|
MTUS1
|
microtubule associated scaffold protein 1
|
|
chrX_+_100666854
Show fit
|
0.11 |
ENST00000640282.1
|
SRPX2
|
sushi repeat containing protein X-linked 2
|
|
chr1_+_205504592
Show fit
|
0.11 |
ENST00000506784.5
ENST00000360066.6
|
CDK18
|
cyclin dependent kinase 18
|
|
chr3_-_58627596
Show fit
|
0.11 |
ENST00000474531.5
ENST00000465970.1
|
FAM107A
|
family with sequence similarity 107 member A
|
|
chr17_+_50426210
Show fit
|
0.11 |
ENST00000506582.5
ENST00000504392.5
ENST00000300441.9
ENST00000427954.6
|
ACSF2
|
acyl-CoA synthetase family member 2
|
|
chr1_+_205504644
Show fit
|
0.11 |
ENST00000429964.7
ENST00000443813.6
|
CDK18
|
cyclin dependent kinase 18
|
|
chr19_-_15934853
Show fit
|
0.10 |
ENST00000620614.4
ENST00000248041.12
|
CYP4F11
|
cytochrome P450 family 4 subfamily F member 11
|
|
chr6_-_43229451
Show fit
|
0.10 |
ENST00000509253.5
ENST00000393987.2
ENST00000230431.11
|
DNPH1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1
|
|
chr3_-_58627567
Show fit
|
0.10 |
ENST00000649301.1
|
FAM107A
|
family with sequence similarity 107 member A
|
|
chr1_-_151826085
Show fit
|
0.10 |
ENST00000356728.11
|
RORC
|
RAR related orphan receptor C
|
|
chr6_-_43059367
Show fit
|
0.09 |
ENST00000230413.9
ENST00000487429.1
ENST00000388752.8
ENST00000489623.1
ENST00000468957.1
|
MRPL2
|
mitochondrial ribosomal protein L2
|
|
chr4_-_158173042
Show fit
|
0.09 |
ENST00000592057.1
ENST00000393807.9
|
GASK1B
|
golgi associated kinase 1B
|
|
chr14_-_99272184
Show fit
|
0.09 |
ENST00000357195.8
|
BCL11B
|
BAF chromatin remodeling complex subunit BCL11B
|
|
chr19_+_15049469
Show fit
|
0.09 |
ENST00000427043.4
|
CASP14
|
caspase 14
|
|
chr7_-_97872420
Show fit
|
0.09 |
ENST00000444334.5
ENST00000422745.5
ENST00000451771.5
ENST00000175506.8
|
ASNS
|
asparagine synthetase (glutamine-hydrolyzing)
|
|
chrX_+_49058966
Show fit
|
0.09 |
ENST00000496529.6
ENST00000606812.5
ENST00000603986.6
ENST00000536628.3
|
CCDC120
|
coiled-coil domain containing 120
|
|
chr1_+_197917355
Show fit
|
0.08 |
ENST00000367388.4
ENST00000367387.6
|
LHX9
|
LIM homeobox 9
|
|
chr16_+_726936
Show fit
|
0.08 |
ENST00000549114.5
ENST00000341413.8
ENST00000562187.1
ENST00000564537.5
ENST00000389703.8
|
HAGHL
|
hydroxyacylglutathione hydrolase like
|
|
chr16_+_727117
Show fit
|
0.08 |
ENST00000562141.5
|
HAGHL
|
hydroxyacylglutathione hydrolase like
|
|
chr18_+_58196736
Show fit
|
0.08 |
ENST00000675221.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase
|
|
chr15_-_21718245
Show fit
|
0.08 |
ENST00000630556.1
|
ENSG00000281179.1
|
novel gene identicle to IGHV1OR15-1
|
|
chr6_+_25652201
Show fit
|
0.08 |
ENST00000612225.4
ENST00000377961.3
|
SCGN
|
secretagogin, EF-hand calcium binding protein
|
|
chr16_+_77722502
Show fit
|
0.08 |
ENST00000564085.5
ENST00000268533.9
ENST00000568787.5
ENST00000437314.3
ENST00000563839.1
|
NUDT7
|
nudix hydrolase 7
|
|
chr3_-_115071333
Show fit
|
0.08 |
ENST00000462705.5
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr1_-_158686700
Show fit
|
0.08 |
ENST00000643759.2
|
SPTA1
|
spectrin alpha, erythrocytic 1
|
|
chr2_+_236569817
Show fit
|
0.08 |
ENST00000272928.4
|
ACKR3
|
atypical chemokine receptor 3
|
|
chr4_-_158173004
Show fit
|
0.08 |
ENST00000585682.6
|
GASK1B
|
golgi associated kinase 1B
|
|
chr2_+_33476640
Show fit
|
0.07 |
ENST00000425210.5
ENST00000444784.5
ENST00000423159.5
ENST00000403687.8
|
RASGRP3
|
RAS guanyl releasing protein 3
|
|
chr7_-_102543849
Show fit
|
0.07 |
ENST00000644544.1
|
UPK3BL2
|
uroplakin 3B like 2
|
|
chr8_-_144416819
Show fit
|
0.07 |
ENST00000526658.1
ENST00000301305.8
|
SLC39A4
|
solute carrier family 39 member 4
|
|
chr9_-_34895722
Show fit
|
0.07 |
ENST00000603640.6
ENST00000603592.1
ENST00000340783.11
|
FAM205C
|
family with sequence similarity 205 member C
|
|
chr14_-_106737547
Show fit
|
0.07 |
ENST00000632209.1
|
IGHV1-69-2
|
immunoglobulin heavy variable 1-69-2
|
|
chr19_-_14979676
Show fit
|
0.07 |
ENST00000598504.5
ENST00000597262.1
|
SLC1A6
|
solute carrier family 1 member 6
|
|
chr8_+_143267435
Show fit
|
0.07 |
ENST00000521682.5
ENST00000340042.2
|
GLI4
|
GLI family zinc finger 4
|
|
chr1_+_220528112
Show fit
|
0.07 |
ENST00000366917.6
ENST00000402574.5
ENST00000611084.4
ENST00000366918.8
|
MARK1
|
microtubule affinity regulating kinase 1
|
|
chr10_-_75235917
Show fit
|
0.07 |
ENST00000469299.1
ENST00000372538.8
|
COMTD1
|
catechol-O-methyltransferase domain containing 1
|
|
chr6_-_159745186
Show fit
|
0.07 |
ENST00000537657.5
|
SOD2
|
superoxide dismutase 2
|
|
chr10_-_49762335
Show fit
|
0.07 |
ENST00000419399.4
ENST00000432695.2
|
OGDHL
|
oxoglutarate dehydrogenase L
|
|
chr5_-_41213505
Show fit
|
0.07 |
ENST00000337836.10
ENST00000433294.1
|
C6
|
complement C6
|
|
chr11_+_111918900
Show fit
|
0.07 |
ENST00000278601.6
|
C11orf52
|
chromosome 11 open reading frame 52
|
|
chr12_+_108562579
Show fit
|
0.07 |
ENST00000311893.14
ENST00000535729.5
ENST00000431221.6
ENST00000547005.5
ENST00000392807.8
ENST00000539593.1
|
ISCU
|
iron-sulfur cluster assembly enzyme
|
|
chr5_+_161850597
Show fit
|
0.07 |
ENST00000634335.1
ENST00000635880.1
|
GABRA1
|
gamma-aminobutyric acid type A receptor subunit alpha1
|
|
chr22_+_22594528
Show fit
|
0.06 |
ENST00000390303.3
|
IGLV3-32
|
immunoglobulin lambda variable 3-32 (non-functional)
|
|
chr1_+_212565334
Show fit
|
0.06 |
ENST00000366981.8
ENST00000366987.6
|
ATF3
|
activating transcription factor 3
|
|
chr22_-_28711931
Show fit
|
0.06 |
ENST00000434810.5
ENST00000456369.5
|
CHEK2
|
checkpoint kinase 2
|
|
chr1_-_161132577
Show fit
|
0.06 |
ENST00000464113.1
|
DEDD
|
death effector domain containing
|
|
chr2_-_25341886
Show fit
|
0.06 |
ENST00000321117.10
|
DNMT3A
|
DNA methyltransferase 3 alpha
|
|
chr1_-_230869564
Show fit
|
0.06 |
ENST00000470540.5
|
C1orf198
|
chromosome 1 open reading frame 198
|
|
chr22_-_28712136
Show fit
|
0.06 |
ENST00000464581.6
|
CHEK2
|
checkpoint kinase 2
|
|
chr5_-_75052546
Show fit
|
0.06 |
ENST00000652361.2
|
GCNT4
|
glucosaminyl (N-acetyl) transferase 4
|
|
chr6_+_31946086
Show fit
|
0.06 |
ENST00000425368.7
|
CFB
|
complement factor B
|
|
chr19_+_48270079
Show fit
|
0.06 |
ENST00000595607.6
|
ZNF114
|
zinc finger protein 114
|
|
chr19_-_53267723
Show fit
|
0.06 |
ENST00000311170.5
|
VN1R4
|
vomeronasal 1 receptor 4
|
|
chr4_-_185810894
Show fit
|
0.06 |
ENST00000448662.6
ENST00000439049.5
ENST00000420158.5
ENST00000319471.13
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr6_-_33580229
Show fit
|
0.06 |
ENST00000374467.4
ENST00000442998.6
ENST00000360661.9
|
BAK1
|
BCL2 antagonist/killer 1
|
|
chr2_+_68774782
Show fit
|
0.06 |
ENST00000409030.7
ENST00000409220.5
|
ARHGAP25
|
Rho GTPase activating protein 25
|
|
chr10_+_122112957
Show fit
|
0.06 |
ENST00000369001.5
|
TACC2
|
transforming acidic coiled-coil containing protein 2
|
|
chr16_+_31117656
Show fit
|
0.06 |
ENST00000219797.9
ENST00000448516.6
|
KAT8
|
lysine acetyltransferase 8
|
|
chr18_+_11751494
Show fit
|
0.05 |
ENST00000269162.9
|
GNAL
|
G protein subunit alpha L
|
|
chr21_+_42219123
Show fit
|
0.05 |
ENST00000398449.8
|
ABCG1
|
ATP binding cassette subfamily G member 1
|
|
chr6_-_25874212
Show fit
|
0.05 |
ENST00000361703.10
ENST00000397060.8
|
SLC17A3
|
solute carrier family 17 member 3
|
|
chr1_-_161132659
Show fit
|
0.05 |
ENST00000368006.8
ENST00000490843.6
ENST00000545495.5
|
DEDD
|
death effector domain containing
|
|
chr15_+_76336755
Show fit
|
0.05 |
ENST00000290759.9
|
ISL2
|
ISL LIM homeobox 2
|
|
chr16_-_370338
Show fit
|
0.05 |
ENST00000450882.1
ENST00000441883.5
ENST00000447696.5
ENST00000389675.6
|
MRPL28
|
mitochondrial ribosomal protein L28
|
|
chr8_-_129786617
Show fit
|
0.05 |
ENST00000276708.9
|
GSDMC
|
gasdermin C
|
|
chr18_+_616711
Show fit
|
0.05 |
ENST00000579494.1
|
CLUL1
|
clusterin like 1
|
|
chr7_-_43729452
Show fit
|
0.05 |
ENST00000310564.10
ENST00000431651.5
ENST00000223336.11
ENST00000415798.5
|
COA1
|
cytochrome c oxidase assembly factor 1 homolog
|
|
chr20_+_3071618
Show fit
|
0.05 |
ENST00000217386.2
|
OXT
|
oxytocin/neurophysin I prepropeptide
|
|
chr9_-_6565462
Show fit
|
0.05 |
ENST00000638661.1
ENST00000640208.1
|
GLDC
|
glycine decarboxylase
|
|
chr19_+_17406099
Show fit
|
0.05 |
ENST00000634942.2
ENST00000634568.1
ENST00000600514.5
|
BISPR
MVB12A
|
BST2 interferon stimulated positive regulator
multivesicular body subunit 12A
|
|
chr18_+_616672
Show fit
|
0.05 |
ENST00000338387.11
|
CLUL1
|
clusterin like 1
|
|
chr12_+_116559381
Show fit
|
0.05 |
ENST00000556529.4
|
MAP1LC3B2
|
microtubule associated protein 1 light chain 3 beta 2
|
|
chr6_+_128883114
Show fit
|
0.05 |
ENST00000421865.3
ENST00000618192.4
ENST00000617695.4
|
LAMA2
|
laminin subunit alpha 2
|
|
chr8_+_143558329
Show fit
|
0.05 |
ENST00000262580.9
ENST00000525721.1
ENST00000534018.5
|
GSDMD
|
gasdermin D
|
|
chr2_-_60553558
Show fit
|
0.05 |
ENST00000642439.1
ENST00000356842.9
|
BCL11A
|
BAF chromatin remodeling complex subunit BCL11A
|
|
chr16_-_48610150
Show fit
|
0.05 |
ENST00000262384.4
|
N4BP1
|
NEDD4 binding protein 1
|
|
chr2_-_68952880
Show fit
|
0.05 |
ENST00000481498.1
ENST00000328895.9
|
GKN2
|
gastrokine 2
|
|
chr1_-_160646958
Show fit
|
0.05 |
ENST00000538290.2
|
SLAMF1
|
signaling lymphocytic activation molecule family member 1
|
|
chr2_-_60553618
Show fit
|
0.05 |
ENST00000643716.1
ENST00000359629.10
ENST00000642384.2
ENST00000335712.11
|
BCL11A
|
BAF chromatin remodeling complex subunit BCL11A
|
|
chr22_+_29767351
Show fit
|
0.05 |
ENST00000330029.6
ENST00000401406.3
|
UQCR10
|
ubiquinol-cytochrome c reductase, complex III subunit X
|
|
chr17_+_18183803
Show fit
|
0.05 |
ENST00000399138.5
|
ALKBH5
|
alkB homolog 5, RNA demethylase
|
|
chr11_+_66546896
Show fit
|
0.05 |
ENST00000513398.2
|
ACTN3
|
actinin alpha 3
|
|
chr6_+_84033717
Show fit
|
0.05 |
ENST00000257776.5
|
MRAP2
|
melanocortin 2 receptor accessory protein 2
|
|
chr9_-_94328644
Show fit
|
0.05 |
ENST00000341207.5
ENST00000253262.9
|
NUTM2F
|
NUT family member 2F
|
|
chr15_-_23687290
Show fit
|
0.05 |
ENST00000649030.2
|
NDN
|
necdin, MAGE family member
|
|
chr1_-_160647287
Show fit
|
0.05 |
ENST00000235739.6
|
SLAMF1
|
signaling lymphocytic activation molecule family member 1
|
|
chr1_+_43979179
Show fit
|
0.05 |
ENST00000434555.7
ENST00000372324.6
ENST00000481924.2
|
B4GALT2
|
beta-1,4-galactosyltransferase 2
|
|
chr2_+_54330867
Show fit
|
0.05 |
ENST00000398634.7
ENST00000405749.5
ENST00000447328.5
ENST00000615983.1
|
C2orf73
|
chromosome 2 open reading frame 73
|
|
chr3_-_167407837
Show fit
|
0.05 |
ENST00000455345.7
|
ZBBX
|
zinc finger B-box domain containing
|
|
chr1_+_110873135
Show fit
|
0.04 |
ENST00000271324.6
|
CD53
|
CD53 molecule
|
|
chr19_-_15898057
Show fit
|
0.04 |
ENST00000011989.11
ENST00000221700.11
|
CYP4F2
|
cytochrome P450 family 4 subfamily F member 2
|
|
chr1_+_19596960
Show fit
|
0.04 |
ENST00000617872.4
ENST00000322753.7
ENST00000602662.1
|
MICOS10
NBL1
|
mitochondrial contact site and cristae organizing system subunit 10
NBL1, DAN family BMP antagonist
|
|
chr14_-_75981986
Show fit
|
0.04 |
ENST00000238682.8
|
TGFB3
|
transforming growth factor beta 3
|
|
chr16_-_370514
Show fit
|
0.04 |
ENST00000199706.13
|
MRPL28
|
mitochondrial ribosomal protein L28
|
|
chr9_-_21187671
Show fit
|
0.04 |
ENST00000421715.2
|
IFNA4
|
interferon alpha 4
|
|
chr5_-_113294895
Show fit
|
0.04 |
ENST00000514701.5
ENST00000302475.8
|
MCC
|
MCC regulator of WNT signaling pathway
|
|
chr11_+_113314811
Show fit
|
0.04 |
ENST00000393020.5
|
TTC12
|
tetratricopeptide repeat domain 12
|
|
chr10_+_122271292
Show fit
|
0.04 |
ENST00000260723.6
|
BTBD16
|
BTB domain containing 16
|
|
chr9_+_128275343
Show fit
|
0.04 |
ENST00000495313.5
ENST00000372898.6
|
SWI5
|
SWI5 homologous recombination repair protein
|
|
chr19_-_344786
Show fit
|
0.04 |
ENST00000264819.7
|
MIER2
|
MIER family member 2
|
|
chr19_+_49496424
Show fit
|
0.04 |
ENST00000596873.1
ENST00000594493.1
ENST00000270625.7
ENST00000599561.1
|
RPS11
|
ribosomal protein S11
|
|
chr14_-_106715166
Show fit
|
0.04 |
ENST00000390633.2
|
IGHV1-69
|
immunoglobulin heavy variable 1-69
|
|
chr8_+_144148640
Show fit
|
0.04 |
ENST00000534366.5
|
MROH1
|
maestro heat like repeat family member 1
|
|
chr9_-_127916978
Show fit
|
0.04 |
ENST00000361444.3
ENST00000335791.10
|
ST6GALNAC4
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 4
|
|
chr11_+_30323077
Show fit
|
0.04 |
ENST00000282032.4
|
ARL14EP
|
ADP ribosylation factor like GTPase 14 effector protein
|
|
chr15_+_74890005
Show fit
|
0.04 |
ENST00000569931.5
ENST00000566377.5
ENST00000569233.5
ENST00000567132.5
ENST00000564633.5
ENST00000568907.5
ENST00000563422.5
ENST00000564003.5
ENST00000562800.5
ENST00000563786.5
ENST00000535694.5
ENST00000323744.10
ENST00000352410.9
ENST00000568828.5
ENST00000562606.5
ENST00000565576.5
ENST00000567570.5
|
MPI
|
mannose phosphate isomerase
|
|
chr12_+_57229694
Show fit
|
0.04 |
ENST00000557487.5
ENST00000328923.8
ENST00000555634.5
ENST00000556689.5
|
SHMT2
|
serine hydroxymethyltransferase 2
|
|
chr18_-_49813512
Show fit
|
0.04 |
ENST00000285093.15
|
ACAA2
|
acetyl-CoA acyltransferase 2
|
|
chr20_+_58689124
Show fit
|
0.04 |
ENST00000525967.5
ENST00000525817.5
|
NPEPL1
|
aminopeptidase like 1
|
|
chr16_-_5097742
Show fit
|
0.04 |
ENST00000587133.1
ENST00000458008.8
ENST00000427587.9
|
EEF2KMT
|
eukaryotic elongation factor 2 lysine methyltransferase
|
|
chr19_+_3762665
Show fit
|
0.04 |
ENST00000330133.5
|
MRPL54
|
mitochondrial ribosomal protein L54
|
|
chr12_+_48328980
Show fit
|
0.04 |
ENST00000335017.1
|
H1-7
|
H1.7 linker histone
|
|
chrX_+_48786578
Show fit
|
0.04 |
ENST00000376670.9
|
GATA1
|
GATA binding protein 1
|
|
chr12_+_56224318
Show fit
|
0.04 |
ENST00000267023.9
ENST00000380198.6
ENST00000341463.5
|
NABP2
|
nucleic acid binding protein 2
|
|
chr5_-_177409535
Show fit
|
0.04 |
ENST00000253496.4
|
F12
|
coagulation factor XII
|
|
chr17_-_28576882
Show fit
|
0.04 |
ENST00000395319.7
ENST00000581807.5
ENST00000226253.9
ENST00000584086.5
ENST00000395321.6
|
ALDOC
|
aldolase, fructose-bisphosphate C
|
|
chr3_-_196318170
Show fit
|
0.04 |
ENST00000325318.10
|
DYNLT2B
|
dynein light chain Tctex-type 2B
|
|
chr1_+_159587817
Show fit
|
0.04 |
ENST00000255040.3
|
APCS
|
amyloid P component, serum
|
|
chr11_+_63974578
Show fit
|
0.04 |
ENST00000314133.4
ENST00000535431.1
|
COX8A
ENSG00000256100.1
|
cytochrome c oxidase subunit 8A
novel transcript
|
|
chr19_-_50476838
Show fit
|
0.04 |
ENST00000600100.6
|
FAM71E1
|
family with sequence similarity 71 member E1
|
|
chr1_+_119414931
Show fit
|
0.04 |
ENST00000543831.5
ENST00000433745.5
ENST00000369416.4
|
HSD3B2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2
|
|
chr14_-_106762576
Show fit
|
0.03 |
ENST00000624687.1
|
IGHV1-69D
|
immunoglobulin heavy variable 1-69D
|
|
chr2_-_86563470
Show fit
|
0.03 |
ENST00000409225.2
|
CHMP3
|
charged multivesicular body protein 3
|
|
chr19_-_11559145
Show fit
|
0.03 |
ENST00000589171.5
ENST00000590700.5
ENST00000586683.5
ENST00000593077.1
ENST00000252445.7
|
ELOF1
|
elongation factor 1 homolog
|
|
chr15_-_26939518
Show fit
|
0.03 |
ENST00000541819.6
|
GABRB3
|
gamma-aminobutyric acid type A receptor subunit beta3
|
|
chr1_-_206921867
Show fit
|
0.03 |
ENST00000628511.2
ENST00000367091.8
|
FCMR
|
Fc fragment of IgM receptor
|
|
chr19_+_55999726
Show fit
|
0.03 |
ENST00000390649.8
ENST00000621651.4
|
NLRP5
|
NLR family pyrin domain containing 5
|
|
chr19_+_3762705
Show fit
|
0.03 |
ENST00000589174.1
|
MRPL54
|
mitochondrial ribosomal protein L54
|
|
chr4_+_127781815
Show fit
|
0.03 |
ENST00000508776.5
|
HSPA4L
|
heat shock protein family A (Hsp70) member 4 like
|
|
chr21_+_42219111
Show fit
|
0.03 |
ENST00000450121.5
ENST00000361802.6
|
ABCG1
|
ATP binding cassette subfamily G member 1
|
|
chr3_-_194351290
Show fit
|
0.03 |
ENST00000429275.1
ENST00000323830.4
|
CPN2
|
carboxypeptidase N subunit 2
|
|
chr8_-_71356653
Show fit
|
0.03 |
ENST00000388742.8
ENST00000388740.4
|
EYA1
|
EYA transcriptional coactivator and phosphatase 1
|
|
chr10_+_79347491
Show fit
|
0.03 |
ENST00000448165.1
|
PPIF
|
peptidylprolyl isomerase F
|
|
chr10_-_118342240
Show fit
|
0.03 |
ENST00000369170.4
|
FAM204A
|
family with sequence similarity 204 member A
|
|
chr7_+_142598016
Show fit
|
0.03 |
ENST00000620773.1
|
TRBV16
|
T cell receptor beta variable 16
|
|
chr19_-_14518383
Show fit
|
0.03 |
ENST00000254322.3
ENST00000595139.2
|
DNAJB1
|
DnaJ heat shock protein family (Hsp40) member B1
|
|
chr11_-_112086727
Show fit
|
0.03 |
ENST00000504148.3
ENST00000541231.1
|
TIMM8B
|
translocase of inner mitochondrial membrane 8 homolog B
|
|
chr19_-_50476725
Show fit
|
0.03 |
ENST00000595790.5
|
FAM71E1
|
family with sequence similarity 71 member E1
|
|
chr14_-_95714088
Show fit
|
0.03 |
ENST00000556450.5
|
TCL1A
|
TCL1 family AKT coactivator A
|
|
chr17_+_7435416
Show fit
|
0.03 |
ENST00000323206.2
ENST00000396568.1
|
TMEM102
|
transmembrane protein 102
|
|
chr2_-_85398734
Show fit
|
0.03 |
ENST00000453973.5
|
CAPG
|
capping actin protein, gelsolin like
|
|
chr3_+_138187511
Show fit
|
0.03 |
ENST00000461822.5
ENST00000485396.5
ENST00000471453.5
ENST00000470821.5
ENST00000471709.5
ENST00000463485.5
|
ARMC8
|
armadillo repeat containing 8
|
|
chr9_+_96928843
Show fit
|
0.03 |
ENST00000372322.4
|
NUTM2G
|
NUT family member 2G
|
|
chr2_-_86563382
Show fit
|
0.03 |
ENST00000263856.9
|
CHMP3
|
charged multivesicular body protein 3
|
|
chr8_-_140635617
Show fit
|
0.03 |
ENST00000220592.10
|
AGO2
|
argonaute RISC catalytic component 2
|
|
chr1_-_202710428
Show fit
|
0.03 |
ENST00000367268.5
|
SYT2
|
synaptotagmin 2
|
|
chr11_-_707063
Show fit
|
0.03 |
ENST00000683307.1
|
DEAF1
|
DEAF1 transcription factor
|
|
chr3_-_146528750
Show fit
|
0.03 |
ENST00000483300.5
|
PLSCR1
|
phospholipid scramblase 1
|
|
chr19_-_49453472
Show fit
|
0.03 |
ENST00000601825.1
ENST00000596049.5
ENST00000599366.5
ENST00000597415.5
|
PIH1D1
|
PIH1 domain containing 1
|
|
chr9_-_38069220
Show fit
|
0.03 |
ENST00000377707.4
|
SHB
|
SH2 domain containing adaptor protein B
|
|
chr19_-_14517425
Show fit
|
0.03 |
ENST00000676577.1
ENST00000677204.1
ENST00000598235.2
|
DNAJB1
|
DnaJ heat shock protein family (Hsp40) member B1
|
|
chr14_-_58427158
Show fit
|
0.03 |
ENST00000555097.1
ENST00000556367.6
ENST00000555404.5
|
TIMM9
|
translocase of inner mitochondrial membrane 9
|
|
chrX_+_48786562
Show fit
|
0.03 |
ENST00000651144.1
ENST00000376665.4
|
GATA1
|
GATA binding protein 1
|
|
chr15_+_41838839
Show fit
|
0.03 |
ENST00000458483.4
|
PLA2G4B
|
phospholipase A2 group IVB
|
|
chr19_-_14529193
Show fit
|
0.03 |
ENST00000596853.6
ENST00000676515.1
ENST00000678338.1
ENST00000595992.6
ENST00000677848.1
ENST00000677762.1
ENST00000678009.1
ENST00000596075.2
ENST00000601533.6
ENST00000396969.8
ENST00000598692.2
ENST00000678098.1
|
DNAJB1
|
DnaJ heat shock protein family (Hsp40) member B1
|
|
chr11_-_62832803
Show fit
|
0.03 |
ENST00000636508.1
|
TEX54
|
testis expressed 54
|
|
chr9_-_21228222
Show fit
|
0.03 |
ENST00000413767.2
|
IFNA17
|
interferon alpha 17
|
|
chr2_-_210225437
Show fit
|
0.03 |
ENST00000233710.4
|
ACADL
|
acyl-CoA dehydrogenase long chain
|
|
chr3_-_139678011
Show fit
|
0.03 |
ENST00000646611.1
ENST00000645290.1
ENST00000647257.1
|
NMNAT3
|
nicotinamide nucleotide adenylyltransferase 3
|
|
chr1_+_40691749
Show fit
|
0.03 |
ENST00000372654.5
|
NFYC
|
nuclear transcription factor Y subunit gamma
|
|
chr4_-_56387422
Show fit
|
0.03 |
ENST00000513376.5
ENST00000205214.11
ENST00000502617.1
ENST00000451613.5
ENST00000602986.5
|
AASDH
|
aminoadipate-semialdehyde dehydrogenase
|
|
chr19_+_38374758
Show fit
|
0.03 |
ENST00000585598.1
ENST00000602911.5
ENST00000592561.5
|
PSMD8
|
proteasome 26S subunit, non-ATPase 8
|
|
chrX_+_22136552
Show fit
|
0.03 |
ENST00000682888.1
ENST00000684356.1
|
PHEX
|
phosphate regulating endopeptidase homolog X-linked
|
|
chr2_+_28630559
Show fit
|
0.03 |
ENST00000436775.1
|
PLB1
|
phospholipase B1
|
|
chr9_-_21385395
Show fit
|
0.03 |
ENST00000380206.4
|
IFNA2
|
interferon alpha 2
|
|
chr7_+_6615576
Show fit
|
0.03 |
ENST00000457543.4
|
ZNF853
|
zinc finger protein 853
|
|
chr14_+_22314435
Show fit
|
0.03 |
ENST00000390467.3
|
TRAV40
|
T cell receptor alpha variable 40
|
|
chrX_+_8464830
Show fit
|
0.03 |
ENST00000453306.4
ENST00000381032.6
ENST00000444481.3
|
VCX3B
|
variable charge X-linked 3B
|
|
chr2_-_86563349
Show fit
|
0.03 |
ENST00000409727.5
|
CHMP3
|
charged multivesicular body protein 3
|
|
chr2_-_174846405
Show fit
|
0.03 |
ENST00000409597.5
ENST00000413882.6
|
CHN1
|
chimerin 1
|
|
chr19_-_43026448
Show fit
|
0.02 |
ENST00000598133.1
ENST00000403486.5
ENST00000320078.12
ENST00000306322.7
|
PSG11
|
pregnancy specific beta-1-glycoprotein 11
|
|
chr20_+_56412249
Show fit
|
0.02 |
ENST00000679887.1
ENST00000434344.2
|
CASS4
|
Cas scaffold protein family member 4
|
|
chr12_+_9827472
Show fit
|
0.02 |
ENST00000617793.4
ENST00000617889.5
ENST00000354855.7
ENST00000279545.7
|
KLRF1
|
killer cell lectin like receptor F1
|
|
chr14_-_58427134
Show fit
|
0.02 |
ENST00000555930.6
|
TIMM9
|
translocase of inner mitochondrial membrane 9
|
|
chr19_+_3933059
Show fit
|
0.02 |
ENST00000616156.4
ENST00000168977.6
ENST00000599576.5
|
NMRK2
|
nicotinamide riboside kinase 2
|