Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
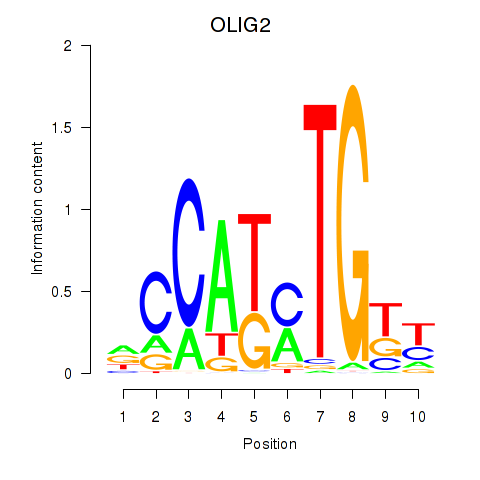
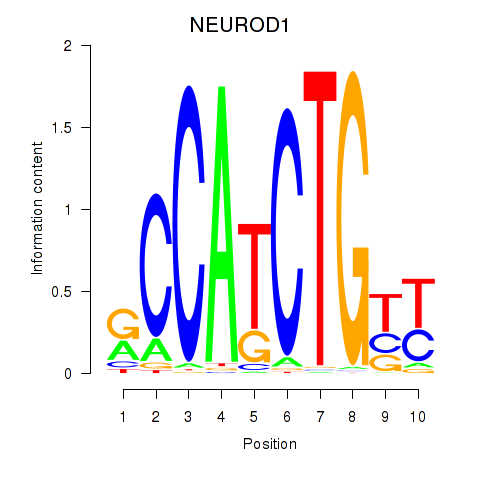
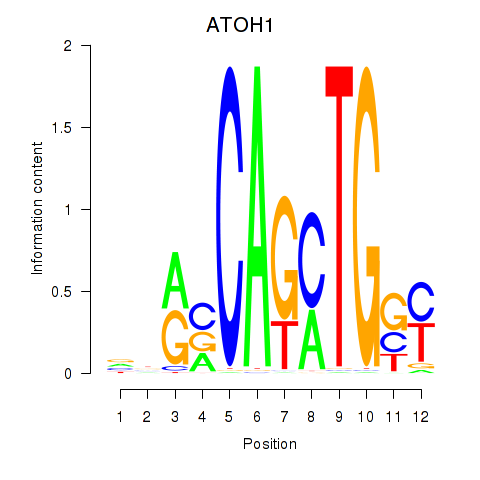
Results for OLIG2_NEUROD1_ATOH1
Z-value: 0.37
Transcription factors associated with OLIG2_NEUROD1_ATOH1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OLIG2
|
ENSG00000205927.5 | oligodendrocyte transcription factor 2 |
|
NEUROD1
|
ENSG00000162992.5 | neuronal differentiation 1 |
|
ATOH1
|
ENSG00000172238.6 | atonal bHLH transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NEUROD1 | hg38_v1_chr2_-_181680490_181680525 | -0.74 | 3.7e-02 | Click! |
| OLIG2 | hg38_v1_chr21_+_33025927_33025942 | 0.31 | 4.5e-01 | Click! |
| ATOH1 | hg38_v1_chr4_+_93828746_93828758 | 0.26 | 5.3e-01 | Click! |
Activity profile of OLIG2_NEUROD1_ATOH1 motif
Sorted Z-values of OLIG2_NEUROD1_ATOH1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_113292838 | 1.05 |
ENST00000672411.1
ENST00000673231.1 |
ANK2
|
ankyrin 2 |
| chr4_+_113292925 | 1.03 |
ENST00000673353.1
ENST00000505342.6 ENST00000672915.1 ENST00000509550.5 |
ANK2
|
ankyrin 2 |
| chr8_-_134510182 | 0.99 |
ENST00000521673.5
|
ZFAT
|
zinc finger and AT-hook domain containing |
| chr4_+_113049616 | 0.76 |
ENST00000504454.5
ENST00000357077.9 ENST00000394537.7 ENST00000672779.1 ENST00000264366.10 |
ANK2
|
ankyrin 2 |
| chr19_-_38253238 | 0.70 |
ENST00000587515.5
|
PPP1R14A
|
protein phosphatase 1 regulatory inhibitor subunit 14A |
| chr12_+_4273751 | 0.66 |
ENST00000675880.1
ENST00000261254.8 |
CCND2
|
cyclin D2 |
| chr4_+_54229261 | 0.42 |
ENST00000508170.5
ENST00000512143.1 ENST00000257290.10 |
PDGFRA
|
platelet derived growth factor receptor alpha |
| chr17_+_41226648 | 0.40 |
ENST00000377721.3
|
KRTAP9-2
|
keratin associated protein 9-2 |
| chr22_+_44677077 | 0.33 |
ENST00000403581.5
|
PRR5
|
proline rich 5 |
| chr22_+_44677044 | 0.31 |
ENST00000006251.11
|
PRR5
|
proline rich 5 |
| chr13_+_87671354 | 0.30 |
ENST00000683689.1
|
SLITRK5
|
SLIT and NTRK like family member 5 |
| chrX_+_136197039 | 0.30 |
ENST00000370683.6
|
FHL1
|
four and a half LIM domains 1 |
| chr16_+_21704963 | 0.30 |
ENST00000388957.3
|
OTOA
|
otoancorin |
| chr19_+_15949008 | 0.30 |
ENST00000322107.1
|
OR10H4
|
olfactory receptor family 10 subfamily H member 4 |
| chr17_-_9791586 | 0.29 |
ENST00000571134.2
|
DHRS7C
|
dehydrogenase/reductase 7C |
| chrX_+_136196750 | 0.29 |
ENST00000539015.5
|
FHL1
|
four and a half LIM domains 1 |
| chr11_+_18602969 | 0.28 |
ENST00000636011.1
ENST00000542172.1 |
SPTY2D1OS
|
SPTY2D1 opposite strand |
| chrX_+_136197020 | 0.28 |
ENST00000370676.7
|
FHL1
|
four and a half LIM domains 1 |
| chr17_-_79952007 | 0.25 |
ENST00000574241.6
|
TBC1D16
|
TBC1 domain family member 16 |
| chr3_-_52452828 | 0.25 |
ENST00000496590.1
|
TNNC1
|
troponin C1, slow skeletal and cardiac type |
| chr19_+_53867874 | 0.25 |
ENST00000448420.5
ENST00000439000.5 ENST00000391771.1 ENST00000391770.9 |
MYADM
|
myeloid associated differentiation marker |
| chr12_+_4269771 | 0.22 |
ENST00000676411.1
|
CCND2
|
cyclin D2 |
| chr15_+_22094522 | 0.21 |
ENST00000328795.5
|
OR4N4
|
olfactory receptor family 4 subfamily N member 4 |
| chr8_-_41665261 | 0.21 |
ENST00000522231.5
ENST00000314214.12 ENST00000348036.8 ENST00000522543.5 |
ANK1
|
ankyrin 1 |
| chr19_-_11339573 | 0.20 |
ENST00000222120.8
|
RAB3D
|
RAB3D, member RAS oncogene family |
| chr10_-_114684457 | 0.20 |
ENST00000392955.7
|
ABLIM1
|
actin binding LIM protein 1 |
| chr10_-_114684612 | 0.20 |
ENST00000533213.6
ENST00000369252.8 |
ABLIM1
|
actin binding LIM protein 1 |
| chr14_+_24115299 | 0.19 |
ENST00000559354.5
ENST00000560459.5 ENST00000559593.5 ENST00000396941.8 ENST00000396936.5 |
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr3_-_18438767 | 0.19 |
ENST00000454909.6
|
SATB1
|
SATB homeobox 1 |
| chr6_+_150143018 | 0.19 |
ENST00000361131.5
|
PPP1R14C
|
protein phosphatase 1 regulatory inhibitor subunit 14C |
| chr11_-_64166102 | 0.19 |
ENST00000255681.7
ENST00000675777.1 |
MACROD1
|
mono-ADP ribosylhydrolase 1 |
| chr20_+_44355692 | 0.19 |
ENST00000316673.8
ENST00000609795.5 ENST00000457232.5 ENST00000609262.5 |
HNF4A
|
hepatocyte nuclear factor 4 alpha |
| chr19_+_40717091 | 0.18 |
ENST00000263370.3
|
ITPKC
|
inositol-trisphosphate 3-kinase C |
| chr17_+_7438267 | 0.18 |
ENST00000575235.5
|
FGF11
|
fibroblast growth factor 11 |
| chr15_+_88638947 | 0.18 |
ENST00000559876.2
|
ISG20
|
interferon stimulated exonuclease gene 20 |
| chr11_+_57598184 | 0.18 |
ENST00000677625.1
ENST00000676670.1 |
SERPING1
|
serpin family G member 1 |
| chr11_+_57597563 | 0.18 |
ENST00000619430.2
ENST00000457869.1 ENST00000340687.10 ENST00000278407.9 ENST00000378323.8 ENST00000378324.6 ENST00000403558.1 |
SERPING1
|
serpin family G member 1 |
| chr7_+_76302665 | 0.18 |
ENST00000248553.7
ENST00000674638.1 ENST00000674547.1 ENST00000675226.1 ENST00000675538.1 ENST00000676231.1 ENST00000675134.1 ENST00000675906.1 ENST00000674650.1 |
HSPB1
|
heat shock protein family B (small) member 1 |
| chr1_-_110064916 | 0.18 |
ENST00000649954.1
|
ALX3
|
ALX homeobox 3 |
| chr5_+_176810552 | 0.16 |
ENST00000329542.9
|
UNC5A
|
unc-5 netrin receptor A |
| chr21_+_29130630 | 0.16 |
ENST00000399926.5
ENST00000399928.6 |
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr12_+_26195313 | 0.16 |
ENST00000422622.3
|
SSPN
|
sarcospan |
| chr19_+_17305801 | 0.16 |
ENST00000602206.1
ENST00000252602.2 |
MRPL34
|
mitochondrial ribosomal protein L34 |
| chr9_+_113536497 | 0.16 |
ENST00000462143.5
|
RGS3
|
regulator of G protein signaling 3 |
| chr12_+_26195543 | 0.15 |
ENST00000242729.7
|
SSPN
|
sarcospan |
| chr5_+_168291599 | 0.15 |
ENST00000265293.9
|
WWC1
|
WW and C2 domain containing 1 |
| chr10_-_60389833 | 0.15 |
ENST00000280772.7
|
ANK3
|
ankyrin 3 |
| chr17_-_58415628 | 0.15 |
ENST00000583753.5
|
RNF43
|
ring finger protein 43 |
| chr1_-_205422050 | 0.15 |
ENST00000367153.9
|
LEMD1
|
LEM domain containing 1 |
| chr7_-_100479142 | 0.15 |
ENST00000300181.7
ENST00000393991.5 |
TSC22D4
|
TSC22 domain family member 4 |
| chr12_+_57455266 | 0.15 |
ENST00000266646.3
|
INHBE
|
inhibin subunit beta E |
| chr7_-_41703062 | 0.15 |
ENST00000242208.5
|
INHBA
|
inhibin subunit beta A |
| chr15_+_88639009 | 0.14 |
ENST00000306072.10
|
ISG20
|
interferon stimulated exonuclease gene 20 |
| chr3_-_100114488 | 0.14 |
ENST00000477258.2
ENST00000354552.7 ENST00000331335.9 ENST00000398326.2 |
FILIP1L
|
filamin A interacting protein 1 like |
| chr19_-_4338786 | 0.14 |
ENST00000601482.1
ENST00000600324.5 ENST00000594605.6 |
STAP2
|
signal transducing adaptor family member 2 |
| chr3_+_119173515 | 0.14 |
ENST00000497685.5
|
UPK1B
|
uroplakin 1B |
| chr4_-_185956652 | 0.14 |
ENST00000355634.9
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr11_-_695215 | 0.14 |
ENST00000382409.4
|
DEAF1
|
DEAF1 transcription factor |
| chr1_+_155078829 | 0.13 |
ENST00000368408.4
|
EFNA3
|
ephrin A3 |
| chr8_-_65789084 | 0.13 |
ENST00000379419.8
|
PDE7A
|
phosphodiesterase 7A |
| chr17_-_3691887 | 0.13 |
ENST00000552050.5
|
P2RX5
|
purinergic receptor P2X 5 |
| chr1_+_172452885 | 0.13 |
ENST00000367725.4
|
C1orf105
|
chromosome 1 open reading frame 105 |
| chr15_+_21651844 | 0.13 |
ENST00000623441.1
|
OR4N4C
|
olfactory receptor family 4 subfamily N member 4C |
| chr17_-_41009124 | 0.13 |
ENST00000391588.3
|
KRTAP3-1
|
keratin associated protein 3-1 |
| chr17_-_41124178 | 0.13 |
ENST00000394014.2
|
KRTAP4-12
|
keratin associated protein 4-12 |
| chr6_-_65707214 | 0.13 |
ENST00000370621.7
ENST00000393380.6 ENST00000503581.6 |
EYS
|
eyes shut homolog |
| chr8_+_106447918 | 0.13 |
ENST00000442977.6
|
OXR1
|
oxidation resistance 1 |
| chr1_-_153550083 | 0.13 |
ENST00000368714.1
|
S100A4
|
S100 calcium binding protein A4 |
| chr10_-_119536533 | 0.12 |
ENST00000392865.5
|
RGS10
|
regulator of G protein signaling 10 |
| chr11_-_119381629 | 0.12 |
ENST00000260187.7
ENST00000455332.6 |
USP2
|
ubiquitin specific peptidase 2 |
| chr7_+_151086466 | 0.12 |
ENST00000397238.7
|
AGAP3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr8_-_92017637 | 0.12 |
ENST00000422361.6
|
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1 |
| chr12_-_56934403 | 0.12 |
ENST00000293502.2
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C member 7 |
| chr4_-_24912886 | 0.12 |
ENST00000504487.5
|
CCDC149
|
coiled-coil domain containing 149 |
| chr8_-_41665200 | 0.12 |
ENST00000335651.6
|
ANK1
|
ankyrin 1 |
| chr10_-_62816341 | 0.12 |
ENST00000242480.4
ENST00000637191.1 |
EGR2
|
early growth response 2 |
| chr19_+_44777860 | 0.12 |
ENST00000341505.4
ENST00000647358.2 |
CBLC
|
Cbl proto-oncogene C |
| chr9_+_12775012 | 0.12 |
ENST00000319264.4
|
LURAP1L
|
leucine rich adaptor protein 1 like |
| chr1_-_150876571 | 0.12 |
ENST00000354396.6
ENST00000358595.10 ENST00000505755.5 |
ARNT
|
aryl hydrocarbon receptor nuclear translocator |
| chr6_+_101393699 | 0.12 |
ENST00000369134.9
ENST00000684068.1 ENST00000683903.1 ENST00000681975.1 |
GRIK2
|
glutamate ionotropic receptor kainate type subunit 2 |
| chr8_-_80080816 | 0.12 |
ENST00000520527.5
ENST00000517427.5 ENST00000379097.7 ENST00000448733.3 |
TPD52
|
tumor protein D52 |
| chr7_-_76626127 | 0.12 |
ENST00000454397.1
|
POMZP3
|
POM121 and ZP3 fusion |
| chr4_-_24912954 | 0.12 |
ENST00000502801.1
ENST00000635206.2 |
CCDC149
|
coiled-coil domain containing 149 |
| chr19_-_4454084 | 0.11 |
ENST00000591919.5
|
UBXN6
|
UBX domain protein 6 |
| chr1_-_150876697 | 0.11 |
ENST00000515192.5
|
ARNT
|
aryl hydrocarbon receptor nuclear translocator |
| chr11_-_89063631 | 0.11 |
ENST00000455756.6
|
GRM5
|
glutamate metabotropic receptor 5 |
| chr7_-_29989774 | 0.11 |
ENST00000242059.10
|
SCRN1
|
secernin 1 |
| chr19_-_6720641 | 0.11 |
ENST00000245907.11
|
C3
|
complement C3 |
| chr7_-_29990113 | 0.11 |
ENST00000426154.5
ENST00000421434.5 ENST00000434476.6 |
SCRN1
|
secernin 1 |
| chr17_-_19748285 | 0.11 |
ENST00000570414.1
ENST00000225740.11 |
ALDH3A1
|
aldehyde dehydrogenase 3 family member A1 |
| chr18_+_13277351 | 0.11 |
ENST00000679091.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr17_-_19748341 | 0.11 |
ENST00000395555.7
|
ALDH3A1
|
aldehyde dehydrogenase 3 family member A1 |
| chr2_+_74002685 | 0.11 |
ENST00000305799.8
|
TET3
|
tet methylcytosine dioxygenase 3 |
| chr17_-_49209217 | 0.11 |
ENST00000515635.5
|
GNGT2
|
G protein subunit gamma transducin 2 |
| chr15_+_40239857 | 0.11 |
ENST00000260404.8
|
PAK6
|
p21 (RAC1) activated kinase 6 |
| chr10_-_62816309 | 0.11 |
ENST00000411732.3
|
EGR2
|
early growth response 2 |
| chr17_-_19748355 | 0.10 |
ENST00000494157.6
|
ALDH3A1
|
aldehyde dehydrogenase 3 family member A1 |
| chr1_+_81306096 | 0.10 |
ENST00000370721.5
ENST00000370727.5 ENST00000370725.5 ENST00000370723.5 ENST00000370728.5 ENST00000370730.5 |
ADGRL2
|
adhesion G protein-coupled receptor L2 |
| chr19_+_35106510 | 0.10 |
ENST00000648240.1
|
ENSG00000285526.1
|
novel protein |
| chr11_-_1486763 | 0.10 |
ENST00000329957.7
|
MOB2
|
MOB kinase activator 2 |
| chr12_-_52473798 | 0.10 |
ENST00000252250.7
|
KRT6C
|
keratin 6C |
| chr14_-_75981986 | 0.10 |
ENST00000238682.8
|
TGFB3
|
transforming growth factor beta 3 |
| chr12_-_7092422 | 0.10 |
ENST00000543835.5
ENST00000647956.2 ENST00000535233.6 |
C1R
|
complement C1r |
| chr11_+_64237420 | 0.10 |
ENST00000541681.1
|
VEGFB
|
vascular endothelial growth factor B |
| chr7_+_74964692 | 0.10 |
ENST00000616305.2
|
CASTOR2
|
cytosolic arginine sensor for mTORC1 subunit 2 |
| chr16_+_29663219 | 0.10 |
ENST00000436527.5
ENST00000360121.4 ENST00000652691.1 ENST00000449759.2 |
SPN
QPRT
|
sialophorin quinolinate phosphoribosyltransferase |
| chr22_+_23894375 | 0.10 |
ENST00000215754.8
|
MIF
|
macrophage migration inhibitory factor |
| chr12_+_26195647 | 0.10 |
ENST00000535504.1
|
SSPN
|
sarcospan |
| chr7_-_122699108 | 0.10 |
ENST00000340112.3
|
RNF133
|
ring finger protein 133 |
| chr21_-_32813679 | 0.10 |
ENST00000487113.1
ENST00000382373.4 |
C21orf62
|
chromosome 21 open reading frame 62 |
| chr19_-_18919348 | 0.09 |
ENST00000349893.8
ENST00000351079.8 ENST00000600932.5 ENST00000262812.9 |
COPE
|
COPI coat complex subunit epsilon |
| chr19_+_35758143 | 0.09 |
ENST00000444637.6
ENST00000396908.8 ENST00000301165.9 |
PROSER3
|
proline and serine rich 3 |
| chr12_+_116559381 | 0.09 |
ENST00000556529.4
|
MAP1LC3B2
|
microtubule associated protein 1 light chain 3 beta 2 |
| chr10_+_100999287 | 0.09 |
ENST00000370220.1
|
LZTS2
|
leucine zipper tumor suppressor 2 |
| chr22_+_44702186 | 0.09 |
ENST00000336985.11
ENST00000403696.5 ENST00000457960.5 ENST00000361473.9 |
PRR5
PRR5-ARHGAP8
|
proline rich 5 PRR5-ARHGAP8 readthrough |
| chr17_-_41118369 | 0.09 |
ENST00000391413.4
|
KRTAP4-11
|
keratin associated protein 4-11 |
| chr8_-_109644766 | 0.09 |
ENST00000533065.5
ENST00000276646.14 |
SYBU
|
syntabulin |
| chr22_+_20917398 | 0.09 |
ENST00000354336.8
|
CRKL
|
CRK like proto-oncogene, adaptor protein |
| chr15_-_65115185 | 0.09 |
ENST00000559089.6
|
UBAP1L
|
ubiquitin associated protein 1 like |
| chr7_+_30134956 | 0.09 |
ENST00000324453.13
ENST00000409688.1 |
MTURN
|
maturin, neural progenitor differentiation regulator homolog |
| chr10_-_96271508 | 0.09 |
ENST00000427367.6
ENST00000413476.6 ENST00000371176.6 |
BLNK
|
B cell linker |
| chr1_-_154870264 | 0.09 |
ENST00000618040.4
ENST00000271915.9 |
KCNN3
|
potassium calcium-activated channel subfamily N member 3 |
| chr10_+_119029711 | 0.09 |
ENST00000425699.3
|
NANOS1
|
nanos C2HC-type zinc finger 1 |
| chrX_+_68693629 | 0.09 |
ENST00000374597.3
|
STARD8
|
StAR related lipid transfer domain containing 8 |
| chr19_+_1241733 | 0.09 |
ENST00000395633.5
ENST00000215375.7 ENST00000591660.5 |
ATP5F1D
|
ATP synthase F1 subunit delta |
| chr20_-_32536383 | 0.09 |
ENST00000375678.7
|
NOL4L
|
nucleolar protein 4 like |
| chr10_-_59709842 | 0.09 |
ENST00000395348.8
|
SLC16A9
|
solute carrier family 16 member 9 |
| chr1_-_225924228 | 0.09 |
ENST00000343818.11
ENST00000612039.4 ENST00000432920.2 |
PYCR2
ENSG00000255835.1
|
pyrroline-5-carboxylate reductase 2 novel protein |
| chr17_-_4987354 | 0.08 |
ENST00000361571.9
|
CAMTA2
|
calmodulin binding transcription activator 2 |
| chr18_+_46333956 | 0.08 |
ENST00000587853.1
|
RNF165
|
ring finger protein 165 |
| chr11_+_117203135 | 0.08 |
ENST00000529622.1
|
TAGLN
|
transgelin |
| chr1_-_74198132 | 0.08 |
ENST00000370911.7
ENST00000370909.6 ENST00000354431.9 |
LRRIQ3
|
leucine rich repeats and IQ motif containing 3 |
| chr2_+_232697362 | 0.08 |
ENST00000482666.5
ENST00000483164.5 ENST00000490229.5 ENST00000464805.5 ENST00000489328.1 |
GIGYF2
|
GRB10 interacting GYF protein 2 |
| chr16_-_84239750 | 0.08 |
ENST00000568181.1
|
KCNG4
|
potassium voltage-gated channel modifier subfamily G member 4 |
| chr10_+_94402486 | 0.08 |
ENST00000225235.5
|
TBC1D12
|
TBC1 domain family member 12 |
| chr11_-_796185 | 0.08 |
ENST00000533385.5
ENST00000528936.5 ENST00000629634.2 ENST00000625752.2 ENST00000528606.5 ENST00000320230.9 |
SLC25A22
|
solute carrier family 25 member 22 |
| chr17_+_51165815 | 0.08 |
ENST00000513177.5
|
NME2
|
NME/NM23 nucleoside diphosphate kinase 2 |
| chr17_-_43661915 | 0.08 |
ENST00000318579.9
ENST00000393661.2 |
MEOX1
|
mesenchyme homeobox 1 |
| chr1_-_153390976 | 0.08 |
ENST00000368732.5
ENST00000368733.4 |
S100A8
|
S100 calcium binding protein A8 |
| chr3_+_189631373 | 0.08 |
ENST00000264731.8
ENST00000418709.6 ENST00000320472.9 ENST00000392460.7 ENST00000440651.6 |
TP63
|
tumor protein p63 |
| chrX_-_49233309 | 0.08 |
ENST00000376251.5
ENST00000323022.10 ENST00000376265.2 |
CACNA1F
|
calcium voltage-gated channel subunit alpha1 F |
| chr10_+_122112957 | 0.08 |
ENST00000369001.5
|
TACC2
|
transforming acidic coiled-coil containing protein 2 |
| chr16_+_21684429 | 0.08 |
ENST00000388956.8
|
OTOA
|
otoancorin |
| chr19_+_46303599 | 0.08 |
ENST00000300862.7
|
HIF3A
|
hypoxia inducible factor 3 subunit alpha |
| chr17_-_49209367 | 0.08 |
ENST00000300406.6
ENST00000511277.5 ENST00000511673.1 |
GNGT2
|
G protein subunit gamma transducin 2 |
| chr2_+_136765508 | 0.08 |
ENST00000409968.6
|
THSD7B
|
thrombospondin type 1 domain containing 7B |
| chr12_+_40692413 | 0.08 |
ENST00000551295.7
ENST00000547702.5 ENST00000551424.5 |
CNTN1
|
contactin 1 |
| chr6_+_26124161 | 0.08 |
ENST00000377791.4
ENST00000602637.1 |
H2AC6
|
H2A clustered histone 6 |
| chr4_-_185956348 | 0.08 |
ENST00000431902.5
ENST00000284776.11 ENST00000415274.5 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr6_+_135181361 | 0.08 |
ENST00000527615.5
ENST00000420123.6 ENST00000525369.5 ENST00000528774.5 ENST00000533624.5 ENST00000534044.5 ENST00000534121.5 |
MYB
|
MYB proto-oncogene, transcription factor |
| chr19_-_49020523 | 0.08 |
ENST00000637680.1
|
ENSG00000268655.2
|
novel protein |
| chr1_+_27935110 | 0.08 |
ENST00000549094.1
|
SMPDL3B
|
sphingomyelin phosphodiesterase acid like 3B |
| chr19_+_15049469 | 0.08 |
ENST00000427043.4
|
CASP14
|
caspase 14 |
| chr11_-_125592448 | 0.08 |
ENST00000648911.1
|
FEZ1
|
fasciculation and elongation protein zeta 1 |
| chr1_-_206772484 | 0.08 |
ENST00000423557.1
|
IL10
|
interleukin 10 |
| chr16_-_79599902 | 0.07 |
ENST00000569649.1
|
MAF
|
MAF bZIP transcription factor |
| chr18_+_58045683 | 0.07 |
ENST00000592846.5
ENST00000675801.1 |
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr1_-_162023632 | 0.07 |
ENST00000367940.2
|
OLFML2B
|
olfactomedin like 2B |
| chr11_-_2149603 | 0.07 |
ENST00000643349.1
|
ENSG00000284779.2
|
novel protein |
| chr2_+_68774782 | 0.07 |
ENST00000409030.7
ENST00000409220.5 |
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr10_-_96271553 | 0.07 |
ENST00000224337.10
|
BLNK
|
B cell linker |
| chr1_+_1632152 | 0.07 |
ENST00000472264.1
ENST00000356026.10 ENST00000378675.7 |
MMP23B
|
matrix metallopeptidase 23B |
| chr7_+_5190273 | 0.07 |
ENST00000382384.6
|
WIPI2
|
WD repeat domain, phosphoinositide interacting 2 |
| chr6_-_30075767 | 0.07 |
ENST00000244360.8
ENST00000376751.8 |
RNF39
|
ring finger protein 39 |
| chr12_-_57752265 | 0.07 |
ENST00000547281.5
ENST00000257904.11 ENST00000546489.5 ENST00000552388.1 |
CDK4
|
cyclin dependent kinase 4 |
| chr1_-_225923891 | 0.07 |
ENST00000472798.2
ENST00000489681.5 ENST00000612651.4 |
PYCR2
|
pyrroline-5-carboxylate reductase 2 |
| chr1_+_6448022 | 0.07 |
ENST00000416731.5
ENST00000461727.6 |
ESPN
|
espin |
| chr12_-_123268077 | 0.07 |
ENST00000542174.5
|
CDK2AP1
|
cyclin dependent kinase 2 associated protein 1 |
| chr5_-_36241798 | 0.07 |
ENST00000514504.5
|
NADK2
|
NAD kinase 2, mitochondrial |
| chr12_-_57752345 | 0.07 |
ENST00000551800.5
ENST00000549606.5 ENST00000312990.10 |
CDK4
|
cyclin dependent kinase 4 |
| chr16_-_88977196 | 0.07 |
ENST00000268679.9
|
CBFA2T3
|
CBFA2/RUNX1 partner transcriptional co-repressor 3 |
| chr7_-_31340678 | 0.07 |
ENST00000297142.4
|
NEUROD6
|
neuronal differentiation 6 |
| chr1_+_27934980 | 0.07 |
ENST00000373894.8
|
SMPDL3B
|
sphingomyelin phosphodiesterase acid like 3B |
| chr10_+_21524627 | 0.07 |
ENST00000651097.1
|
MLLT10
|
MLLT10 histone lysine methyltransferase DOT1L cofactor |
| chr9_+_132162045 | 0.07 |
ENST00000393229.4
|
NTNG2
|
netrin G2 |
| chr17_+_40177460 | 0.07 |
ENST00000620260.6
|
RAPGEFL1
|
Rap guanine nucleotide exchange factor like 1 |
| chr12_+_112978424 | 0.07 |
ENST00000680685.1
ENST00000620097.2 |
OAS2
|
2'-5'-oligoadenylate synthetase 2 |
| chr10_+_49610297 | 0.07 |
ENST00000374115.5
|
SLC18A3
|
solute carrier family 18 member A3 |
| chr15_-_63156774 | 0.07 |
ENST00000462430.5
|
RPS27L
|
ribosomal protein S27 like |
| chr12_+_112978460 | 0.07 |
ENST00000449768.2
|
OAS2
|
2'-5'-oligoadenylate synthetase 2 |
| chr11_-_95232514 | 0.07 |
ENST00000634898.1
ENST00000542176.1 ENST00000278499.6 |
SESN3
|
sestrin 3 |
| chr4_-_185812209 | 0.07 |
ENST00000393523.6
ENST00000393528.7 ENST00000449407.6 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr18_+_3448456 | 0.07 |
ENST00000549780.5
|
TGIF1
|
TGFB induced factor homeobox 1 |
| chrX_+_118974608 | 0.07 |
ENST00000304778.11
ENST00000371628.8 |
LONRF3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr2_+_190649062 | 0.07 |
ENST00000409581.5
ENST00000337386.10 |
NAB1
|
NGFI-A binding protein 1 |
| chr1_-_115841116 | 0.07 |
ENST00000320238.3
|
NHLH2
|
nescient helix-loop-helix 2 |
| chr7_-_11832190 | 0.07 |
ENST00000423059.9
ENST00000617773.1 |
THSD7A
|
thrombospondin type 1 domain containing 7A |
| chr15_+_66453418 | 0.07 |
ENST00000566326.1
|
MAP2K1
|
mitogen-activated protein kinase kinase 1 |
| chr16_+_23835946 | 0.07 |
ENST00000321728.12
ENST00000643927.1 |
PRKCB
|
protein kinase C beta |
| chr2_+_127423265 | 0.07 |
ENST00000402125.2
|
PROC
|
protein C, inactivator of coagulation factors Va and VIIIa |
| chr17_+_69502397 | 0.06 |
ENST00000613873.4
ENST00000589647.5 |
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr18_+_58045642 | 0.06 |
ENST00000676223.1
ENST00000675147.1 |
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr11_-_64245816 | 0.06 |
ENST00000542235.1
|
PPP1R14B
|
protein phosphatase 1 regulatory inhibitor subunit 14B |
| chr8_-_22156789 | 0.06 |
ENST00000306317.7
|
LGI3
|
leucine rich repeat LGI family member 3 |
| chr8_-_140764386 | 0.06 |
ENST00000520151.5
ENST00000519024.5 ENST00000519465.5 |
PTK2
|
protein tyrosine kinase 2 |
| chr2_+_127419882 | 0.06 |
ENST00000409048.1
|
PROC
|
protein C, inactivator of coagulation factors Va and VIIIa |
| chr6_+_42746958 | 0.06 |
ENST00000614467.4
|
BICRAL
|
BRD4 interacting chromatin remodeling complex associated protein like |
| chr7_+_129368123 | 0.06 |
ENST00000460109.5
ENST00000474594.5 |
AHCYL2
|
adenosylhomocysteinase like 2 |
| chr2_+_184598520 | 0.06 |
ENST00000302277.7
|
ZNF804A
|
zinc finger protein 804A |
| chr3_+_100114911 | 0.06 |
ENST00000489081.5
|
CMSS1
|
cms1 ribosomal small subunit homolog |
Network of associatons between targets according to the STRING database.
First level regulatory network of OLIG2_NEUROD1_ATOH1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.8 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.4 | GO:0072275 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.1 | 0.4 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.1 | 0.3 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.1 | 0.2 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.3 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 0.2 | GO:0021593 | rhombomere morphogenesis(GO:0021593) rhombomere 3 morphogenesis(GO:0021658) |
| 0.0 | 0.8 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.2 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.1 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.9 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.1 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.0 | 0.2 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.2 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.1 | GO:0002445 | type IIa hypersensitivity(GO:0001794) regulation of type IIa hypersensitivity(GO:0001796) type II hypersensitivity(GO:0002445) regulation of type II hypersensitivity(GO:0002892) |
| 0.0 | 0.1 | GO:0044725 | chromatin reprogramming in the zygote(GO:0044725) |
| 0.0 | 0.1 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 | 0.2 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.0 | 0.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.1 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.9 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.1 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) negative regulation of hypersensitivity(GO:0002884) |
| 0.0 | 0.1 | GO:1904636 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.0 | 0.2 | GO:0046886 | positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.0 | 0.1 | GO:0060809 | mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 0.0 | 0.1 | GO:1904899 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.0 | 0.1 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.2 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.1 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 | 0.2 | GO:0038031 | non-canonical Wnt signaling pathway via MAPK cascade(GO:0038030) non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.2 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0043449 | olfactory learning(GO:0008355) cellular alkene metabolic process(GO:0043449) |
| 0.0 | 0.1 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.0 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.0 | 0.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.1 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.0 | 0.1 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.0 | 0.2 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.4 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:1905247 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.0 | 0.2 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.1 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.0 | 0.1 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.0 | 0.1 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) interleukin-3-mediated signaling pathway(GO:0038156) |
| 0.0 | 0.1 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.0 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.0 | GO:0061182 | negative regulation of chondrocyte development(GO:0061182) |
| 0.0 | 0.1 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.2 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0015870 | acetylcholine transport(GO:0015870) |
| 0.0 | 0.0 | GO:0033869 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) |
| 0.0 | 0.0 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.9 | GO:0003254 | regulation of membrane depolarization(GO:0003254) |
| 0.0 | 0.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.0 | 0.1 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.0 | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 0.0 | 0.0 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.3 | GO:0021756 | striatum development(GO:0021756) |
| 0.0 | 0.1 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 2.9 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.3 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.8 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.4 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.3 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.0 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.3 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.1 | 0.2 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.1 | 0.3 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 3.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.2 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.3 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.0 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.2 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.1 | GO:0016497 | substance K receptor activity(GO:0016497) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.1 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.1 | GO:0004914 | interleukin-3 receptor activity(GO:0004912) interleukin-5 receptor activity(GO:0004914) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.1 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.4 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.0 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.1 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.6 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 1.0 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.3 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |