Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for OLIG3_NEUROD2_NEUROG2
Z-value: 1.94
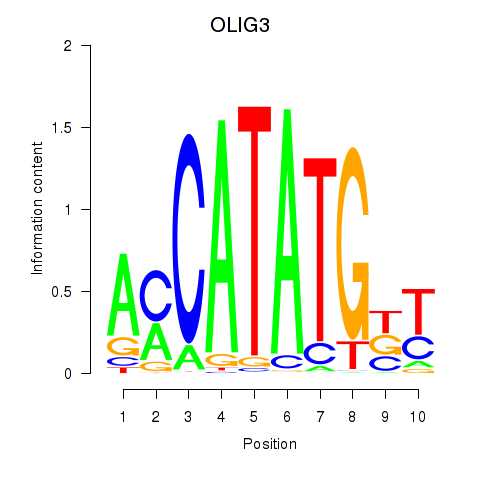
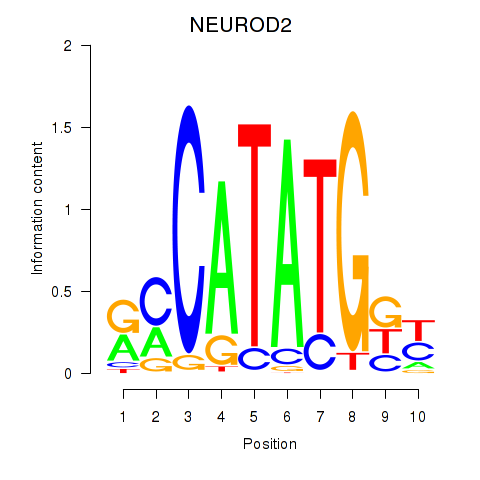
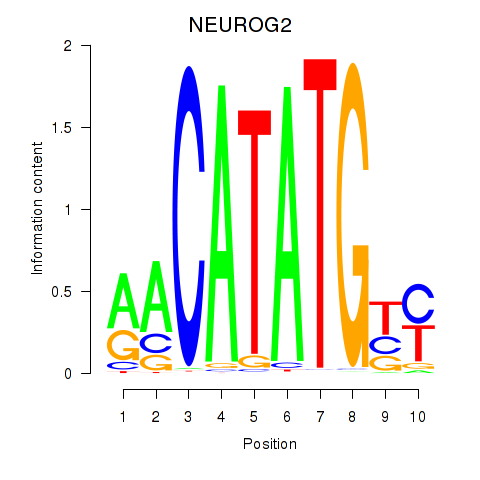
Transcription factors associated with OLIG3_NEUROD2_NEUROG2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OLIG3
|
ENSG00000177468.7 | oligodendrocyte transcription factor 3 |
|
NEUROD2
|
ENSG00000171532.5 | neuronal differentiation 2 |
|
NEUROG2
|
ENSG00000178403.4 | neurogenin 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NEUROD2 | hg38_v1_chr17_-_39607876_39607966 | -0.68 | 6.3e-02 | Click! |
| NEUROG2 | hg38_v1_chr4_-_112516176_112516181 | 0.63 | 9.7e-02 | Click! |
| OLIG3 | hg38_v1_chr6_-_137494387_137494400 | -0.31 | 4.5e-01 | Click! |
Activity profile of OLIG3_NEUROD2_NEUROG2 motif
Sorted Z-values of OLIG3_NEUROD2_NEUROG2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_91179355 | 3.68 |
ENST00000550563.5
ENST00000546370.5 |
DCN
|
decorin |
| chr5_-_111976925 | 3.58 |
ENST00000395634.7
|
NREP
|
neuronal regeneration related protein |
| chr12_-_91179472 | 3.37 |
ENST00000550099.5
ENST00000546391.5 |
DCN
|
decorin |
| chr12_-_91153149 | 2.33 |
ENST00000550758.1
|
DCN
|
decorin |
| chr2_-_187513641 | 2.24 |
ENST00000392365.5
ENST00000435414.5 |
TFPI
|
tissue factor pathway inhibitor |
| chr3_-_195583931 | 1.94 |
ENST00000343267.8
ENST00000421243.5 ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr2_+_188974364 | 1.89 |
ENST00000304636.9
ENST00000317840.9 |
COL3A1
|
collagen type III alpha 1 chain |
| chr8_-_92017637 | 1.53 |
ENST00000422361.6
|
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1 |
| chr5_-_76623391 | 1.44 |
ENST00000296641.5
ENST00000504899.1 |
F2RL2
|
coagulation factor II thrombin receptor like 2 |
| chr9_+_18474100 | 1.14 |
ENST00000327883.11
ENST00000431052.6 ENST00000380570.8 ENST00000380548.9 |
ADAMTSL1
|
ADAMTS like 1 |
| chr15_-_63156774 | 1.04 |
ENST00000462430.5
|
RPS27L
|
ribosomal protein S27 like |
| chr4_+_113292838 | 0.95 |
ENST00000672411.1
ENST00000673231.1 |
ANK2
|
ankyrin 2 |
| chr6_-_56851888 | 0.94 |
ENST00000312431.10
ENST00000520645.5 |
DST
|
dystonin |
| chr4_+_113292925 | 0.93 |
ENST00000673353.1
ENST00000505342.6 ENST00000672915.1 ENST00000509550.5 |
ANK2
|
ankyrin 2 |
| chr6_+_72366730 | 0.93 |
ENST00000414192.2
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr9_-_92482499 | 0.91 |
ENST00000375544.7
|
ASPN
|
asporin |
| chr12_-_108320635 | 0.89 |
ENST00000412676.5
ENST00000550573.5 |
CMKLR1
|
chemerin chemokine-like receptor 1 |
| chr1_-_186375671 | 0.88 |
ENST00000451586.1
|
TPR
|
translocated promoter region, nuclear basket protein |
| chr3_-_114758940 | 0.85 |
ENST00000464560.5
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr3_-_114759115 | 0.82 |
ENST00000471418.5
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr16_+_21704963 | 0.81 |
ENST00000388957.3
|
OTOA
|
otoancorin |
| chr16_+_53099100 | 0.80 |
ENST00000565832.5
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr20_+_36214373 | 0.79 |
ENST00000432603.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1 like 1 |
| chr2_+_157257687 | 0.78 |
ENST00000259056.5
|
GALNT5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr2_+_209580024 | 0.77 |
ENST00000392194.5
|
MAP2
|
microtubule associated protein 2 |
| chr6_-_46491431 | 0.72 |
ENST00000371374.6
|
RCAN2
|
regulator of calcineurin 2 |
| chr4_+_54229261 | 0.71 |
ENST00000508170.5
ENST00000512143.1 ENST00000257290.10 |
PDGFRA
|
platelet derived growth factor receptor alpha |
| chr1_+_103750406 | 0.71 |
ENST00000370079.3
|
AMY1C
|
amylase alpha 1C |
| chr3_-_18438767 | 0.69 |
ENST00000454909.6
|
SATB1
|
SATB homeobox 1 |
| chr2_+_209579598 | 0.68 |
ENST00000445941.5
ENST00000673860.1 |
MAP2
|
microtubule associated protein 2 |
| chr6_-_31680377 | 0.68 |
ENST00000383237.4
|
LY6G5C
|
lymphocyte antigen 6 family member G5C |
| chr4_-_158173004 | 0.66 |
ENST00000585682.6
|
GASK1B
|
golgi associated kinase 1B |
| chr7_-_98252117 | 0.66 |
ENST00000420697.1
ENST00000415086.5 ENST00000447648.7 |
TECPR1
|
tectonin beta-propeller repeat containing 1 |
| chr6_-_46491956 | 0.66 |
ENST00000306764.11
|
RCAN2
|
regulator of calcineurin 2 |
| chr22_-_28306645 | 0.66 |
ENST00000612946.4
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr7_+_134745460 | 0.65 |
ENST00000436461.6
|
CALD1
|
caldesmon 1 |
| chr12_+_25973748 | 0.62 |
ENST00000542865.5
|
RASSF8
|
Ras association domain family member 8 |
| chr4_+_159267737 | 0.62 |
ENST00000264431.8
|
RAPGEF2
|
Rap guanine nucleotide exchange factor 2 |
| chr2_+_33134579 | 0.62 |
ENST00000418533.6
|
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr16_-_66549839 | 0.62 |
ENST00000527800.6
ENST00000677555.1 ENST00000563369.6 |
TK2
|
thymidine kinase 2 |
| chr10_-_77090722 | 0.61 |
ENST00000638531.1
|
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr3_-_9952337 | 0.61 |
ENST00000411976.2
ENST00000412055.6 |
PRRT3
|
proline rich transmembrane protein 3 |
| chr2_+_33134620 | 0.60 |
ENST00000402934.5
ENST00000404525.5 ENST00000407925.5 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr5_-_59430600 | 0.60 |
ENST00000636120.1
|
PDE4D
|
phosphodiesterase 4D |
| chr14_-_74612226 | 0.59 |
ENST00000261978.9
|
LTBP2
|
latent transforming growth factor beta binding protein 2 |
| chr22_+_35381086 | 0.58 |
ENST00000216117.9
ENST00000677931.1 ENST00000679074.1 |
HMOX1
|
heme oxygenase 1 |
| chr2_-_162243375 | 0.57 |
ENST00000188790.9
ENST00000443424.5 |
FAP
|
fibroblast activation protein alpha |
| chr1_+_89524819 | 0.54 |
ENST00000439853.6
ENST00000330947.7 ENST00000449440.5 ENST00000640258.1 |
LRRC8B
|
leucine rich repeat containing 8 VRAC subunit B |
| chr1_+_89524871 | 0.53 |
ENST00000639264.1
|
LRRC8B
|
leucine rich repeat containing 8 VRAC subunit B |
| chr2_-_174598206 | 0.53 |
ENST00000392546.6
ENST00000436221.1 |
WIPF1
|
WAS/WASL interacting protein family member 1 |
| chr6_-_52994248 | 0.52 |
ENST00000457564.1
ENST00000370960.5 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chr3_-_179266971 | 0.50 |
ENST00000349697.2
ENST00000497599.5 |
KCNMB3
|
potassium calcium-activated channel subfamily M regulatory beta subunit 3 |
| chr2_+_108377947 | 0.49 |
ENST00000272452.7
|
SULT1C4
|
sulfotransferase family 1C member 4 |
| chr20_-_41317602 | 0.49 |
ENST00000559234.5
ENST00000683867.1 |
ZHX3
|
zinc fingers and homeoboxes 3 |
| chr4_-_158173042 | 0.47 |
ENST00000592057.1
ENST00000393807.9 |
GASK1B
|
golgi associated kinase 1B |
| chr7_+_29563820 | 0.47 |
ENST00000319694.3
|
PRR15
|
proline rich 15 |
| chr12_-_91179517 | 0.45 |
ENST00000551354.1
|
DCN
|
decorin |
| chr6_+_2999885 | 0.45 |
ENST00000397717.7
ENST00000380455.11 |
NQO2
|
N-ribosyldihydronicotinamide:quinone reductase 2 |
| chr6_+_2999961 | 0.44 |
ENST00000338130.7
|
NQO2
|
N-ribosyldihydronicotinamide:quinone reductase 2 |
| chr6_+_2999984 | 0.44 |
ENST00000380441.5
ENST00000380454.8 |
NQO2
|
N-ribosyldihydronicotinamide:quinone reductase 2 |
| chr1_-_53738024 | 0.43 |
ENST00000628545.1
|
GLIS1
|
GLIS family zinc finger 1 |
| chr12_+_54854505 | 0.43 |
ENST00000308796.11
ENST00000619042.1 |
MUCL1
|
mucin like 1 |
| chr18_-_58629084 | 0.42 |
ENST00000361673.4
|
ALPK2
|
alpha kinase 2 |
| chr15_+_62561361 | 0.42 |
ENST00000561311.5
|
TLN2
|
talin 2 |
| chrX_+_71578435 | 0.41 |
ENST00000373696.8
|
GCNA
|
germ cell nuclear acidic peptidase |
| chr8_-_134510182 | 0.41 |
ENST00000521673.5
|
ZFAT
|
zinc finger and AT-hook domain containing |
| chr15_-_55408018 | 0.40 |
ENST00000569205.5
|
CCPG1
|
cell cycle progression 1 |
| chr17_+_14301069 | 0.39 |
ENST00000360954.3
|
HS3ST3B1
|
heparan sulfate-glucosamine 3-sulfotransferase 3B1 |
| chr17_-_55732074 | 0.39 |
ENST00000575734.5
|
TMEM100
|
transmembrane protein 100 |
| chr3_+_148739798 | 0.39 |
ENST00000402260.2
|
AGTR1
|
angiotensin II receptor type 1 |
| chr10_+_134465 | 0.39 |
ENST00000439456.5
ENST00000397962.8 ENST00000309776.8 ENST00000397959.7 |
ZMYND11
|
zinc finger MYND-type containing 11 |
| chr9_+_2110354 | 0.39 |
ENST00000634772.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr2_+_135741717 | 0.39 |
ENST00000415164.5
|
UBXN4
|
UBX domain protein 4 |
| chr22_+_43923755 | 0.38 |
ENST00000423180.2
ENST00000216180.8 |
PNPLA3
|
patatin like phospholipase domain containing 3 |
| chr6_-_2744126 | 0.38 |
ENST00000647417.1
|
MYLK4
|
myosin light chain kinase family member 4 |
| chr2_-_181680490 | 0.36 |
ENST00000684145.1
ENST00000295108.4 ENST00000684079.1 ENST00000683430.1 |
CERKL
NEUROD1
|
ceramide kinase like neuronal differentiation 1 |
| chr2_+_108378176 | 0.36 |
ENST00000409309.3
|
SULT1C4
|
sulfotransferase family 1C member 4 |
| chr1_-_19484635 | 0.34 |
ENST00000433834.5
|
CAPZB
|
capping actin protein of muscle Z-line subunit beta |
| chr5_-_172006567 | 0.34 |
ENST00000517395.6
ENST00000265094.9 ENST00000393802.6 |
FBXW11
|
F-box and WD repeat domain containing 11 |
| chr10_-_28282086 | 0.34 |
ENST00000375719.7
ENST00000375732.5 |
MPP7
|
membrane palmitoylated protein 7 |
| chr17_+_68515399 | 0.34 |
ENST00000588188.6
|
PRKAR1A
|
protein kinase cAMP-dependent type I regulatory subunit alpha |
| chr3_+_141324208 | 0.34 |
ENST00000509842.5
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr5_+_141359970 | 0.34 |
ENST00000522605.2
ENST00000622527.1 |
PCDHGB2
|
protocadherin gamma subfamily B, 2 |
| chr2_+_10368645 | 0.33 |
ENST00000613496.4
|
HPCAL1
|
hippocalcin like 1 |
| chr9_-_127873462 | 0.33 |
ENST00000223836.10
|
AK1
|
adenylate kinase 1 |
| chr7_-_38363476 | 0.33 |
ENST00000426402.2
|
TRGV2
|
T cell receptor gamma variable 2 |
| chr17_-_50767505 | 0.32 |
ENST00000450727.6
|
ANKRD40CL
|
ANKRD40 C-terminal like |
| chr5_-_138338325 | 0.32 |
ENST00000510119.1
ENST00000513970.5 |
CDC25C
|
cell division cycle 25C |
| chr10_-_48604952 | 0.31 |
ENST00000417912.6
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr19_+_14440254 | 0.31 |
ENST00000342216.8
|
PKN1
|
protein kinase N1 |
| chr9_+_76459152 | 0.30 |
ENST00000444201.6
ENST00000376730.5 |
GCNT1
|
glucosaminyl (N-acetyl) transferase 1 |
| chr3_-_149377637 | 0.30 |
ENST00000305366.8
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr13_-_113658197 | 0.30 |
ENST00000335288.5
|
ATP4B
|
ATPase H+/K+ transporting subunit beta |
| chr16_-_4846196 | 0.30 |
ENST00000589389.5
|
GLYR1
|
glyoxylate reductase 1 homolog |
| chr12_+_133080875 | 0.29 |
ENST00000412146.6
ENST00000544426.5 ENST00000355557.7 ENST00000319849.7 ENST00000440550.6 |
ZNF140
|
zinc finger protein 140 |
| chr8_-_23457677 | 0.29 |
ENST00000356206.10
ENST00000417069.6 |
ENTPD4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr5_+_136160986 | 0.29 |
ENST00000507637.1
|
SMAD5
|
SMAD family member 5 |
| chr7_+_148590760 | 0.29 |
ENST00000307003.3
|
C7orf33
|
chromosome 7 open reading frame 33 |
| chr9_-_120477354 | 0.28 |
ENST00000416449.5
|
CDK5RAP2
|
CDK5 regulatory subunit associated protein 2 |
| chr12_-_10454485 | 0.27 |
ENST00000408006.7
ENST00000544822.2 ENST00000536188.5 |
KLRC1
|
killer cell lectin like receptor C1 |
| chr5_-_172006817 | 0.27 |
ENST00000296933.10
|
FBXW11
|
F-box and WD repeat domain containing 11 |
| chr1_+_148889403 | 0.27 |
ENST00000464103.5
ENST00000534536.5 ENST00000369356.8 ENST00000369354.7 ENST00000369347.8 ENST00000369349.7 ENST00000369351.7 |
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr13_+_31945826 | 0.26 |
ENST00000647500.1
|
FRY
|
FRY microtubule binding protein |
| chr11_-_26722051 | 0.26 |
ENST00000396005.8
|
SLC5A12
|
solute carrier family 5 member 12 |
| chr21_+_36135071 | 0.26 |
ENST00000290354.6
|
CBR3
|
carbonyl reductase 3 |
| chr14_+_22147988 | 0.25 |
ENST00000390457.2
|
TRAV27
|
T cell receptor alpha variable 27 |
| chr2_+_71068278 | 0.25 |
ENST00000613852.4
ENST00000455662.6 ENST00000531934.5 |
NAGK
|
N-acetylglucosamine kinase |
| chr9_+_12775012 | 0.25 |
ENST00000319264.4
|
LURAP1L
|
leucine rich adaptor protein 1 like |
| chr2_-_165203870 | 0.25 |
ENST00000639244.1
ENST00000409101.7 ENST00000668657.1 |
SCN3A
|
sodium voltage-gated channel alpha subunit 3 |
| chr5_+_141968886 | 0.25 |
ENST00000347642.7
|
RNF14
|
ring finger protein 14 |
| chr9_+_132162045 | 0.25 |
ENST00000393229.4
|
NTNG2
|
netrin G2 |
| chr12_-_130839230 | 0.25 |
ENST00000392373.7
ENST00000261653.10 |
STX2
|
syntaxin 2 |
| chr2_+_10368764 | 0.24 |
ENST00000620771.4
|
HPCAL1
|
hippocalcin like 1 |
| chr4_+_70592253 | 0.24 |
ENST00000322937.10
ENST00000613447.4 |
AMBN
|
ameloblastin |
| chr1_-_111427731 | 0.24 |
ENST00000369732.4
|
OVGP1
|
oviductal glycoprotein 1 |
| chr10_-_48605032 | 0.24 |
ENST00000249601.9
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr15_+_74173693 | 0.23 |
ENST00000249842.8
|
ISLR
|
immunoglobulin superfamily containing leucine rich repeat |
| chr8_-_23457618 | 0.23 |
ENST00000358689.9
ENST00000518718.1 |
ENTPD4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chrX_-_103502853 | 0.23 |
ENST00000372633.1
|
RAB40A
|
RAB40A, member RAS oncogene family |
| chr10_-_13707536 | 0.23 |
ENST00000632570.1
ENST00000477221.2 |
FRMD4A
|
FERM domain containing 4A |
| chr5_+_141373878 | 0.22 |
ENST00000517434.3
ENST00000610583.1 |
PCDHGA6
|
protocadherin gamma subfamily A, 6 |
| chr9_-_101594995 | 0.21 |
ENST00000636434.1
|
PPP3R2
|
protein phosphatase 3 regulatory subunit B, beta |
| chr2_-_177392673 | 0.21 |
ENST00000447413.1
ENST00000397057.6 ENST00000456746.5 ENST00000464747.5 |
ENSG00000213963.6
NFE2L2
|
novel transcript nuclear factor, erythroid 2 like 2 |
| chrX_-_70289888 | 0.21 |
ENST00000239666.9
ENST00000374454.1 |
PDZD11
|
PDZ domain containing 11 |
| chr17_-_8867639 | 0.21 |
ENST00000619866.5
|
PIK3R6
|
phosphoinositide-3-kinase regulatory subunit 6 |
| chrX_+_47585212 | 0.21 |
ENST00000445623.1
|
TIMP1
|
TIMP metallopeptidase inhibitor 1 |
| chr22_+_42074240 | 0.21 |
ENST00000321753.8
|
PHETA2
|
PH domain containing endocytic trafficking adaptor 2 |
| chr16_+_28878382 | 0.21 |
ENST00000357084.7
|
ATP2A1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr2_+_74554692 | 0.20 |
ENST00000233668.10
ENST00000340004.6 |
DOK1
|
docking protein 1 |
| chr20_-_35529618 | 0.20 |
ENST00000246199.5
ENST00000424444.1 ENST00000374345.8 ENST00000444723.3 |
C20orf173
|
chromosome 20 open reading frame 173 |
| chr2_-_97094882 | 0.20 |
ENST00000414820.6
ENST00000272610.3 |
FAHD2B
|
fumarylacetoacetate hydrolase domain containing 2B |
| chr11_+_77066985 | 0.19 |
ENST00000456580.6
|
CAPN5
|
calpain 5 |
| chr7_+_91264426 | 0.19 |
ENST00000287934.4
|
FZD1
|
frizzled class receptor 1 |
| chr17_-_15260752 | 0.19 |
ENST00000676329.1
ENST00000675551.1 ENST00000644020.1 ENST00000674947.1 |
PMP22
|
peripheral myelin protein 22 |
| chr1_+_248508073 | 0.19 |
ENST00000641804.1
|
OR2G6
|
olfactory receptor family 2 subfamily G member 6 |
| chr11_+_77066948 | 0.19 |
ENST00000527066.5
ENST00000648180.1 ENST00000529629.5 |
CAPN5
|
calpain 5 |
| chr12_+_9971402 | 0.19 |
ENST00000304361.9
ENST00000396507.7 ENST00000434319.6 |
CLEC12A
|
C-type lectin domain family 12 member A |
| chr2_+_54115437 | 0.19 |
ENST00000303536.8
ENST00000394666.7 |
ACYP2
|
acylphosphatase 2 |
| chr16_+_28878480 | 0.19 |
ENST00000395503.9
|
ATP2A1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr19_+_11925062 | 0.19 |
ENST00000622593.4
ENST00000590798.1 |
ZNF700
ENSG00000267179.1
|
zinc finger protein 700 novel protein |
| chr17_-_69268812 | 0.18 |
ENST00000586811.1
|
ABCA5
|
ATP binding cassette subfamily A member 5 |
| chrX_-_13817346 | 0.18 |
ENST00000356942.9
|
GPM6B
|
glycoprotein M6B |
| chr11_+_124183219 | 0.18 |
ENST00000641351.2
|
OR10D3
|
olfactory receptor family 10 subfamily D member 3 |
| chr19_+_11925098 | 0.18 |
ENST00000254321.10
ENST00000591944.1 |
ZNF700
ENSG00000267179.1
|
zinc finger protein 700 novel protein |
| chr11_-_60855943 | 0.18 |
ENST00000332539.5
|
PTGDR2
|
prostaglandin D2 receptor 2 |
| chr11_-_134253248 | 0.18 |
ENST00000392595.6
ENST00000352327.5 ENST00000341541.8 ENST00000392594.7 |
THYN1
|
thymocyte nuclear protein 1 |
| chr19_+_44259903 | 0.18 |
ENST00000588489.5
ENST00000391958.6 |
ZNF233
|
zinc finger protein 233 |
| chr3_-_16513643 | 0.18 |
ENST00000334133.9
|
RFTN1
|
raftlin, lipid raft linker 1 |
| chr1_-_155241220 | 0.17 |
ENST00000368373.8
ENST00000427500.7 |
GBA
|
glucosylceramidase beta |
| chr12_-_10849464 | 0.17 |
ENST00000544994.5
ENST00000228811.8 ENST00000540107.2 |
PRR4
|
proline rich 4 |
| chr11_-_89063631 | 0.17 |
ENST00000455756.6
|
GRM5
|
glutamate metabotropic receptor 5 |
| chr12_+_56468561 | 0.17 |
ENST00000338146.7
|
SPRYD4
|
SPRY domain containing 4 |
| chr6_-_111793871 | 0.17 |
ENST00000368667.6
|
FYN
|
FYN proto-oncogene, Src family tyrosine kinase |
| chrX_+_118974608 | 0.17 |
ENST00000304778.11
ENST00000371628.8 |
LONRF3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr21_-_42366525 | 0.17 |
ENST00000291527.3
|
TFF1
|
trefoil factor 1 |
| chr19_-_21329400 | 0.17 |
ENST00000356929.3
|
ZNF708
|
zinc finger protein 708 |
| chr11_+_134253531 | 0.17 |
ENST00000374752.6
ENST00000281182.9 |
ACAD8
|
acyl-CoA dehydrogenase family member 8 |
| chr6_+_69232406 | 0.17 |
ENST00000238918.12
|
ADGRB3
|
adhesion G protein-coupled receptor B3 |
| chr17_-_60392333 | 0.17 |
ENST00000590133.5
|
USP32
|
ubiquitin specific peptidase 32 |
| chr5_+_141370236 | 0.17 |
ENST00000576222.2
ENST00000618934.1 |
PCDHGB3
|
protocadherin gamma subfamily B, 3 |
| chr3_-_146460440 | 0.16 |
ENST00000610787.5
|
PLSCR2
|
phospholipid scramblase 2 |
| chr4_+_73853290 | 0.16 |
ENST00000226524.4
|
PF4V1
|
platelet factor 4 variant 1 |
| chr19_+_55640966 | 0.16 |
ENST00000590190.1
ENST00000325333.10 ENST00000585995.1 ENST00000592996.5 |
ZNF580
ZNF581
CCDC106
|
zinc finger protein 580 zinc finger protein 581 coiled-coil domain containing 106 |
| chr5_+_141192330 | 0.16 |
ENST00000239446.6
|
PCDHB10
|
protocadherin beta 10 |
| chr21_-_30480364 | 0.16 |
ENST00000390689.3
|
KRTAP19-1
|
keratin associated protein 19-1 |
| chr19_+_8413270 | 0.16 |
ENST00000381035.8
ENST00000595142.5 ENST00000601724.5 ENST00000601283.5 ENST00000215555.7 ENST00000595213.1 |
MARCHF2
|
membrane associated ring-CH-type finger 2 |
| chr6_+_151239951 | 0.16 |
ENST00000402676.7
|
AKAP12
|
A-kinase anchoring protein 12 |
| chr9_+_121299793 | 0.16 |
ENST00000373818.8
|
GSN
|
gelsolin |
| chr15_-_50265666 | 0.16 |
ENST00000543581.5
ENST00000267845.8 |
HDC
|
histidine decarboxylase |
| chr5_+_149357999 | 0.16 |
ENST00000274569.9
|
PCYOX1L
|
prenylcysteine oxidase 1 like |
| chr4_-_88823165 | 0.16 |
ENST00000508369.5
|
FAM13A
|
family with sequence similarity 13 member A |
| chr17_+_19648915 | 0.15 |
ENST00000672567.1
ENST00000672709.1 |
ALDH3A2
|
aldehyde dehydrogenase 3 family member A2 |
| chr16_-_30894264 | 0.15 |
ENST00000380317.8
|
BCL7C
|
BAF chromatin remodeling complex subunit BCL7C |
| chr2_+_201129318 | 0.15 |
ENST00000417748.1
|
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chr10_-_48251757 | 0.15 |
ENST00000305531.3
|
FRMPD2
|
FERM and PDZ domain containing 2 |
| chr3_-_126357399 | 0.15 |
ENST00000296233.4
|
KLF15
|
Kruppel like factor 15 |
| chr5_+_170861990 | 0.15 |
ENST00000523189.6
|
RANBP17
|
RAN binding protein 17 |
| chr9_+_107306459 | 0.15 |
ENST00000457811.1
|
RAD23B
|
RAD23 homolog B, nucleotide excision repair protein |
| chr17_-_44268119 | 0.15 |
ENST00000399246.3
ENST00000262418.12 |
SLC4A1
|
solute carrier family 4 member 1 (Diego blood group) |
| chr3_-_53882142 | 0.15 |
ENST00000335754.8
|
ACTR8
|
actin related protein 8 |
| chr9_-_124941054 | 0.15 |
ENST00000373555.9
|
GOLGA1
|
golgin A1 |
| chr14_-_23435652 | 0.14 |
ENST00000355349.4
|
MYH7
|
myosin heavy chain 7 |
| chr6_+_41042557 | 0.14 |
ENST00000373158.6
ENST00000470917.1 |
TSPO2
|
translocator protein 2 |
| chrX_+_152914426 | 0.14 |
ENST00000318504.11
ENST00000449285.6 ENST00000539731.5 ENST00000535861.5 ENST00000370268.8 ENST00000370270.6 |
ZNF185
|
zinc finger protein 185 with LIM domain |
| chr3_-_146528750 | 0.14 |
ENST00000483300.5
|
PLSCR1
|
phospholipid scramblase 1 |
| chr1_+_100345018 | 0.14 |
ENST00000635056.2
ENST00000647005.1 |
CDC14A
|
cell division cycle 14A |
| chr2_+_177392734 | 0.14 |
ENST00000680770.1
ENST00000637633.2 ENST00000679459.1 ENST00000409888.1 ENST00000264167.11 ENST00000642466.2 |
AGPS
|
alkylglycerone phosphate synthase |
| chr9_+_110048598 | 0.14 |
ENST00000434623.6
ENST00000374525.5 |
PALM2AKAP2
|
PALM2 and AKAP2 fusion |
| chrX_+_154542194 | 0.14 |
ENST00000618670.4
|
IKBKG
|
inhibitor of nuclear factor kappa B kinase regulatory subunit gamma |
| chr4_-_7871986 | 0.14 |
ENST00000360265.9
|
AFAP1
|
actin filament associated protein 1 |
| chr3_-_49429252 | 0.14 |
ENST00000615713.4
|
NICN1
|
nicolin 1 |
| chr16_+_53054973 | 0.14 |
ENST00000447540.6
ENST00000615216.4 ENST00000566029.5 |
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr9_+_128920966 | 0.14 |
ENST00000428610.5
ENST00000372592.8 |
PHYHD1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr12_+_133130618 | 0.13 |
ENST00000426665.6
ENST00000248211.11 |
ZNF10
|
zinc finger protein 10 |
| chrX_+_70290077 | 0.13 |
ENST00000374403.4
|
KIF4A
|
kinesin family member 4A |
| chr4_-_88823214 | 0.13 |
ENST00000513837.5
ENST00000503556.5 |
FAM13A
|
family with sequence similarity 13 member A |
| chr2_-_105396943 | 0.13 |
ENST00000409807.5
|
FHL2
|
four and a half LIM domains 2 |
| chr5_-_131797030 | 0.13 |
ENST00000615660.4
|
FNIP1
|
folliculin interacting protein 1 |
| chr5_-_134174765 | 0.13 |
ENST00000520417.1
|
SKP1
|
S-phase kinase associated protein 1 |
| chr7_+_100049765 | 0.13 |
ENST00000456748.6
ENST00000292450.9 ENST00000438937.1 ENST00000543588.2 |
ZSCAN21
|
zinc finger and SCAN domain containing 21 |
Network of associatons between targets according to the STRING database.
First level regulatory network of OLIG3_NEUROD2_NEUROG2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.6 | 9.8 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.3 | 0.9 | GO:0031453 | regulation of heterochromatin assembly(GO:0031445) positive regulation of heterochromatin assembly(GO:0031453) |
| 0.3 | 1.9 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.2 | 2.2 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.2 | 0.7 | GO:0072277 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.2 | 0.6 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.2 | 0.6 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.2 | 1.9 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.2 | 0.5 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 0.2 | 0.6 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.2 | 1.2 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.2 | 0.9 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 0.6 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.5 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.1 | 0.6 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.1 | 0.4 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.1 | 0.4 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.3 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.1 | 1.2 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.1 | 0.3 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.1 | 0.2 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.1 | 0.2 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 0.4 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 0.2 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.1 | 0.2 | GO:1904457 | beta-glucoside metabolic process(GO:1901804) beta-glucoside catabolic process(GO:1901805) positive regulation of neuronal action potential(GO:1904457) |
| 0.1 | 1.5 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 0.9 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 0.2 | GO:1901377 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.1 | 0.2 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.1 | 0.2 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.1 | 0.5 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.5 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 0.3 | GO:0042376 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.0 | 0.3 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 0.3 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.5 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.0 | 0.3 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.3 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.2 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.8 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 1.2 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.0 | 0.2 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.2 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.0 | 0.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.2 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.0 | 1.0 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.3 | GO:0006050 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.0 | 0.1 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.0 | 0.9 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.2 | GO:1903173 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.0 | 0.9 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 | 0.1 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.1 | GO:0019049 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.8 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.0 | 0.3 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.0 | 0.1 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.4 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.1 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.0 | 0.2 | GO:0044858 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.1 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 0.0 | 0.4 | GO:0036155 | acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.0 | 0.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.1 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.2 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.4 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.1 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 1.5 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.2 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 1.6 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.3 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 1.3 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.1 | GO:0036215 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) diapedesis(GO:0050904) |
| 0.0 | 0.6 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.1 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.2 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.3 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.1 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.0 | 0.1 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.1 | GO:0043323 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.0 | 0.2 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.5 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.0 | 0.3 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.1 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.0 | 0.1 | GO:0010607 | negative regulation of cytoplasmic mRNA processing body assembly(GO:0010607) |
| 0.0 | 0.3 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.1 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 | 0.1 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.3 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.1 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 | 0.1 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.0 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.2 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.0 | 0.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.1 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.0 | 0.4 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.0 | 0.1 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.6 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.1 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.0 | 0.1 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.2 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.3 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.1 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.0 | 0.1 | GO:0051257 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.0 | 0.1 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.1 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.0 | GO:0050992 | dimethylallyl diphosphate biosynthetic process(GO:0050992) dimethylallyl diphosphate metabolic process(GO:0050993) |
| 0.0 | 0.8 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.7 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.7 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.1 | GO:2000580 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 1.5 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.0 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.1 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.6 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.0 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.0 | 0.2 | GO:2000680 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.0 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.0 | 0.1 | GO:0071033 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.0 | 0.1 | GO:0010918 | positive regulation of mitochondrial membrane potential(GO:0010918) |
| 0.0 | 0.1 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.0 | GO:1903182 | regulation of SUMO transferase activity(GO:1903182) positive regulation of SUMO transferase activity(GO:1903755) |
| 0.0 | 0.2 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 9.8 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.2 | 1.5 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.2 | 0.5 | GO:0097636 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.2 | 0.9 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 1.9 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.2 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.1 | 0.2 | GO:0097180 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.1 | 1.3 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.8 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.2 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.9 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.6 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.0 | 2.1 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.2 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.1 | GO:0097229 | sperm end piece(GO:0097229) |
| 0.0 | 0.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.3 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.8 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 2.9 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 0.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 3.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.2 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.0 | GO:1990356 | sumoylated E2 ligase complex(GO:1990356) |
| 0.0 | 1.2 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.3 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0001512 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 0.2 | 0.7 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.2 | 1.2 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.2 | 1.5 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.2 | 1.9 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 1.4 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.4 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 0.6 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 9.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.3 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.1 | 0.3 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.1 | 0.6 | GO:0004797 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.1 | 0.5 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 0.3 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.1 | 0.3 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 0.3 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.1 | 1.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 0.3 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.1 | 0.6 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 0.2 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.1 | 0.4 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.4 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 0.3 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 0.2 | GO:0046577 | long-chain-alcohol oxidase activity(GO:0046577) |
| 0.0 | 0.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.8 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 1.0 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 1.0 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.3 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.6 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.2 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.0 | 0.2 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.1 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.1 | GO:0044594 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.0 | 0.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.1 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 2.1 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0052836 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 0.1 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.0 | 0.4 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 0.6 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.0 | 0.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.3 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.2 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.5 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.3 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.1 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.1 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.1 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.6 | GO:0004950 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 0.5 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.0 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.0 | 0.1 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0098960 | postsynaptic neurotransmitter receptor activity(GO:0098960) neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.4 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 2.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.0 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.0 | 0.0 | GO:0009032 | thymidine phosphorylase activity(GO:0009032) pyrimidine-nucleoside phosphorylase activity(GO:0016154) |
| 0.0 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 10.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.5 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 2.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.7 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.0 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.5 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.9 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 0.7 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 2.2 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.9 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 1.5 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 2.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.7 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.9 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 1.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |