Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
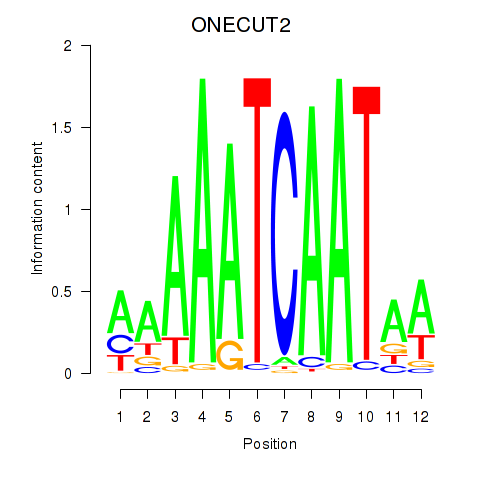
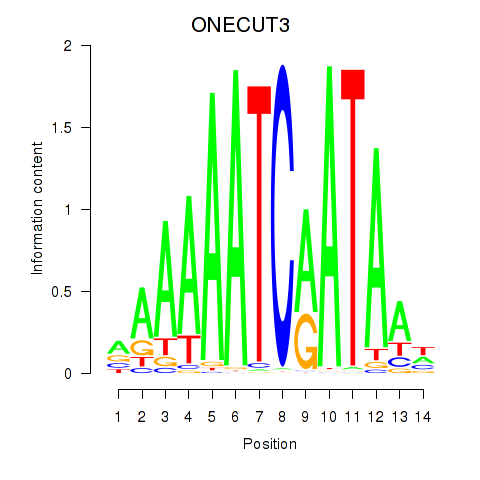
Results for ONECUT2_ONECUT3
Z-value: 0.09
Transcription factors associated with ONECUT2_ONECUT3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ONECUT2
|
ENSG00000119547.6 | one cut homeobox 2 |
|
ONECUT3
|
ENSG00000205922.5 | one cut homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ONECUT2 | hg38_v1_chr18_+_57435366_57435391 | 0.58 | 1.4e-01 | Click! |
| ONECUT3 | hg38_v1_chr19_+_1753499_1753521 | 0.43 | 2.9e-01 | Click! |
Activity profile of ONECUT2_ONECUT3 motif
Sorted Z-values of ONECUT2_ONECUT3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_107498041 | 0.15 |
ENST00000297450.7
|
ANGPT1
|
angiopoietin 1 |
| chr17_+_17782108 | 0.15 |
ENST00000395774.1
|
RAI1
|
retinoic acid induced 1 |
| chr8_-_107497909 | 0.14 |
ENST00000517746.6
|
ANGPT1
|
angiopoietin 1 |
| chr3_+_159839847 | 0.11 |
ENST00000445224.6
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr12_-_91153149 | 0.11 |
ENST00000550758.1
|
DCN
|
decorin |
| chr4_+_70592295 | 0.10 |
ENST00000449493.2
|
AMBN
|
ameloblastin |
| chr1_-_115841116 | 0.10 |
ENST00000320238.3
|
NHLH2
|
nescient helix-loop-helix 2 |
| chr17_+_7558296 | 0.10 |
ENST00000438470.5
ENST00000436057.5 |
TNFSF13
|
TNF superfamily member 13 |
| chr7_-_151633182 | 0.09 |
ENST00000476632.2
|
PRKAG2
|
protein kinase AMP-activated non-catalytic subunit gamma 2 |
| chr12_+_6724071 | 0.08 |
ENST00000229251.7
ENST00000539735.5 ENST00000538410.5 |
COPS7A
|
COP9 signalosome subunit 7A |
| chr12_+_6724008 | 0.08 |
ENST00000626119.2
ENST00000543155.6 |
COPS7A
|
COP9 signalosome subunit 7A |
| chr7_-_23470469 | 0.08 |
ENST00000258729.8
|
IGF2BP3
|
insulin like growth factor 2 mRNA binding protein 3 |
| chr1_-_686673 | 0.08 |
ENST00000332831.4
|
OR4F16
|
olfactory receptor family 4 subfamily F member 16 |
| chr1_+_159587817 | 0.08 |
ENST00000255040.3
|
APCS
|
amyloid P component, serum |
| chr5_+_181367268 | 0.07 |
ENST00000456475.1
|
OR4F3
|
olfactory receptor family 4 subfamily F member 3 |
| chr3_-_142028597 | 0.07 |
ENST00000467667.5
|
TFDP2
|
transcription factor Dp-2 |
| chrX_-_64976500 | 0.07 |
ENST00000447788.6
|
ZC4H2
|
zinc finger C4H2-type containing |
| chrX_-_64976435 | 0.07 |
ENST00000374839.8
|
ZC4H2
|
zinc finger C4H2-type containing |
| chr8_-_167024 | 0.07 |
ENST00000320901.4
|
OR4F21
|
olfactory receptor family 4 subfamily F member 21 |
| chr14_+_36657560 | 0.07 |
ENST00000402703.6
|
PAX9
|
paired box 9 |
| chr10_-_125816596 | 0.06 |
ENST00000368786.5
|
UROS
|
uroporphyrinogen III synthase |
| chr1_-_243163310 | 0.06 |
ENST00000492145.1
ENST00000490813.5 ENST00000464936.5 |
CEP170
|
centrosomal protein 170 |
| chr3_+_69936583 | 0.06 |
ENST00000314557.10
ENST00000394351.9 |
MITF
|
melanocyte inducing transcription factor |
| chr10_+_68109433 | 0.06 |
ENST00000613327.4
ENST00000358913.10 ENST00000373675.3 |
MYPN
|
myopalladin |
| chr13_-_35855627 | 0.06 |
ENST00000379893.5
|
DCLK1
|
doublecortin like kinase 1 |
| chr8_-_144529048 | 0.06 |
ENST00000527462.1
ENST00000313465.5 ENST00000524821.6 |
C8orf82
|
chromosome 8 open reading frame 82 |
| chr14_+_22543179 | 0.06 |
ENST00000390534.1
|
TRAJ3
|
T cell receptor alpha joining 3 |
| chr3_-_142028617 | 0.06 |
ENST00000477292.5
ENST00000478006.5 ENST00000495310.5 ENST00000486111.5 |
TFDP2
|
transcription factor Dp-2 |
| chr16_-_8868343 | 0.06 |
ENST00000562843.5
ENST00000561530.5 ENST00000396593.6 |
CARHSP1
|
calcium regulated heat stable protein 1 |
| chr14_-_22957100 | 0.06 |
ENST00000555367.5
|
HAUS4
|
HAUS augmin like complex subunit 4 |
| chr7_+_129368123 | 0.06 |
ENST00000460109.5
ENST00000474594.5 |
AHCYL2
|
adenosylhomocysteinase like 2 |
| chr14_-_22957061 | 0.06 |
ENST00000557591.5
ENST00000541587.6 ENST00000490506.5 ENST00000554406.1 |
HAUS4
|
HAUS augmin like complex subunit 4 |
| chr14_-_22957128 | 0.06 |
ENST00000342454.12
ENST00000555986.5 ENST00000554516.5 ENST00000347758.6 ENST00000206474.11 ENST00000555040.5 |
HAUS4
|
HAUS augmin like complex subunit 4 |
| chr19_+_37507129 | 0.06 |
ENST00000586138.5
ENST00000588578.5 ENST00000587986.5 |
ZNF793
|
zinc finger protein 793 |
| chr1_-_53220589 | 0.06 |
ENST00000294360.5
|
CZIB
|
CXXC motif containing zinc binding protein |
| chr12_+_4590075 | 0.06 |
ENST00000540757.6
|
DYRK4
|
dual specificity tyrosine phosphorylation regulated kinase 4 |
| chr3_-_24165548 | 0.06 |
ENST00000280696.9
|
THRB
|
thyroid hormone receptor beta |
| chr5_+_139561159 | 0.06 |
ENST00000505007.5
|
UBE2D2
|
ubiquitin conjugating enzyme E2 D2 |
| chr4_+_174918355 | 0.05 |
ENST00000505141.5
ENST00000359240.7 ENST00000615367.4 ENST00000445694.5 ENST00000618444.1 |
ADAM29
|
ADAM metallopeptidase domain 29 |
| chr4_+_174918400 | 0.05 |
ENST00000404450.8
ENST00000514159.1 |
ADAM29
|
ADAM metallopeptidase domain 29 |
| chr2_-_191151568 | 0.05 |
ENST00000358470.8
ENST00000432798.1 ENST00000450994.1 |
STAT4
|
signal transducer and activator of transcription 4 |
| chr1_-_17439657 | 0.05 |
ENST00000375436.9
|
RCC2
|
regulator of chromosome condensation 2 |
| chr12_-_76423256 | 0.05 |
ENST00000546946.5
|
OSBPL8
|
oxysterol binding protein like 8 |
| chr3_+_109136707 | 0.05 |
ENST00000622536.6
|
C3orf85
|
chromosome 3 open reading frame 85 |
| chr7_-_28180735 | 0.05 |
ENST00000283928.10
|
JAZF1
|
JAZF zinc finger 1 |
| chr16_-_8868687 | 0.05 |
ENST00000618335.4
ENST00000570125.5 |
CARHSP1
|
calcium regulated heat stable protein 1 |
| chr20_-_1994046 | 0.05 |
ENST00000217305.3
ENST00000650874.1 |
PDYN
|
prodynorphin |
| chr12_-_54259531 | 0.05 |
ENST00000550411.5
ENST00000439541.6 |
CBX5
|
chromobox 5 |
| chr7_-_105269007 | 0.05 |
ENST00000357311.7
|
SRPK2
|
SRSF protein kinase 2 |
| chr4_-_69961007 | 0.04 |
ENST00000353151.3
|
CSN2
|
casein beta |
| chr5_+_139561308 | 0.04 |
ENST00000398733.8
|
UBE2D2
|
ubiquitin conjugating enzyme E2 D2 |
| chr11_+_63369779 | 0.04 |
ENST00000279178.4
|
SLC22A9
|
solute carrier family 22 member 9 |
| chr4_+_70050431 | 0.04 |
ENST00000511674.5
ENST00000246896.8 |
HTN1
|
histatin 1 |
| chr19_-_44448435 | 0.04 |
ENST00000588655.1
ENST00000592308.1 ENST00000614049.5 ENST00000613197.4 |
ENSG00000267188.1
ZNF229
|
novel transcript zinc finger protein 229 |
| chrX_-_120559889 | 0.04 |
ENST00000371323.3
|
CUL4B
|
cullin 4B |
| chr6_-_154430495 | 0.04 |
ENST00000424998.3
|
CNKSR3
|
CNKSR family member 3 |
| chr11_+_126327863 | 0.04 |
ENST00000648516.1
|
DCPS
|
decapping enzyme, scavenger |
| chr7_+_43112593 | 0.04 |
ENST00000453890.5
ENST00000395891.7 |
HECW1
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 |
| chr20_-_22585451 | 0.04 |
ENST00000377115.4
|
FOXA2
|
forkhead box A2 |
| chr1_+_206557157 | 0.04 |
ENST00000577571.5
|
RASSF5
|
Ras association domain family member 5 |
| chr17_-_352784 | 0.04 |
ENST00000577079.5
ENST00000331302.12 ENST00000618002.4 ENST00000536489.6 |
RPH3AL
|
rabphilin 3A like (without C2 domains) |
| chr3_+_63443076 | 0.04 |
ENST00000295894.9
|
SYNPR
|
synaptoporin |
| chr3_+_40100007 | 0.04 |
ENST00000539167.2
|
MYRIP
|
myosin VIIA and Rab interacting protein |
| chr17_-_68955332 | 0.04 |
ENST00000269080.6
ENST00000615593.4 ENST00000586539.6 ENST00000430352.6 |
ABCA8
|
ATP binding cassette subfamily A member 8 |
| chr10_-_96271508 | 0.04 |
ENST00000427367.6
ENST00000413476.6 ENST00000371176.6 |
BLNK
|
B cell linker |
| chr3_-_187670385 | 0.03 |
ENST00000287641.4
|
SST
|
somatostatin |
| chr1_-_29230871 | 0.03 |
ENST00000373791.7
ENST00000263702.11 |
MECR
|
mitochondrial trans-2-enoyl-CoA reductase |
| chr16_-_20691256 | 0.03 |
ENST00000307493.8
|
ACSM1
|
acyl-CoA synthetase medium chain family member 1 |
| chr5_-_147453888 | 0.03 |
ENST00000398514.7
|
DPYSL3
|
dihydropyrimidinase like 3 |
| chr1_+_207089233 | 0.03 |
ENST00000243611.9
ENST00000367076.7 |
C4BPB
|
complement component 4 binding protein beta |
| chr1_+_207089283 | 0.03 |
ENST00000391923.1
|
C4BPB
|
complement component 4 binding protein beta |
| chr19_+_926001 | 0.03 |
ENST00000263620.8
|
ARID3A
|
AT-rich interaction domain 3A |
| chr1_-_231421146 | 0.03 |
ENST00000667629.1
ENST00000670301.1 ENST00000658954.1 |
EGLN1
|
egl-9 family hypoxia inducible factor 1 |
| chr3_-_71581540 | 0.03 |
ENST00000650068.1
|
FOXP1
|
forkhead box P1 |
| chr10_-_96271553 | 0.03 |
ENST00000224337.10
|
BLNK
|
B cell linker |
| chr1_-_451678 | 0.03 |
ENST00000426406.4
|
OR4F29
|
olfactory receptor family 4 subfamily F member 29 |
| chr9_-_14307928 | 0.03 |
ENST00000637640.1
ENST00000493697.1 ENST00000636057.1 |
NFIB
|
nuclear factor I B |
| chr3_-_123980727 | 0.03 |
ENST00000620893.4
|
ROPN1
|
rhophilin associated tail protein 1 |
| chr20_+_37777262 | 0.03 |
ENST00000373469.1
|
CTNNBL1
|
catenin beta like 1 |
| chr9_-_92482350 | 0.03 |
ENST00000375543.2
|
ASPN
|
asporin |
| chr7_-_100641507 | 0.03 |
ENST00000431692.5
ENST00000223051.8 |
TFR2
|
transferrin receptor 2 |
| chr10_-_119536533 | 0.03 |
ENST00000392865.5
|
RGS10
|
regulator of G protein signaling 10 |
| chr16_-_66918839 | 0.03 |
ENST00000565235.2
ENST00000568632.5 ENST00000565796.5 |
CDH16
|
cadherin 16 |
| chr9_-_135961310 | 0.03 |
ENST00000371756.4
|
UBAC1
|
UBA domain containing 1 |
| chr22_-_35824373 | 0.03 |
ENST00000473487.6
|
RBFOX2
|
RNA binding fox-1 homolog 2 |
| chr11_-_35419213 | 0.03 |
ENST00000642171.1
ENST00000644050.1 ENST00000643134.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr1_+_207089195 | 0.03 |
ENST00000452902.6
|
C4BPB
|
complement component 4 binding protein beta |
| chr3_-_64225436 | 0.02 |
ENST00000638394.2
|
PRICKLE2
|
prickle planar cell polarity protein 2 |
| chr16_-_66918876 | 0.02 |
ENST00000570262.5
ENST00000299752.9 ENST00000394055.7 |
CDH16
|
cadherin 16 |
| chr11_+_8019193 | 0.02 |
ENST00000534099.5
|
TUB
|
TUB bipartite transcription factor |
| chr2_-_49974083 | 0.02 |
ENST00000636345.1
|
NRXN1
|
neurexin 1 |
| chr2_-_49973939 | 0.02 |
ENST00000630656.1
|
NRXN1
|
neurexin 1 |
| chr11_-_58844484 | 0.02 |
ENST00000532258.1
|
GLYATL2
|
glycine-N-acyltransferase like 2 |
| chr19_+_9087061 | 0.02 |
ENST00000641627.1
|
OR1M1
|
olfactory receptor family 1 subfamily M member 1 |
| chr2_+_108588286 | 0.02 |
ENST00000332345.10
|
LIMS1
|
LIM zinc finger domain containing 1 |
| chr11_-_35418966 | 0.02 |
ENST00000531628.2
|
SLC1A2
|
solute carrier family 1 member 2 |
| chr6_+_34236865 | 0.02 |
ENST00000674029.1
ENST00000447654.5 ENST00000347617.10 ENST00000401473.7 ENST00000311487.9 |
HMGA1
|
high mobility group AT-hook 1 |
| chr8_+_67952028 | 0.02 |
ENST00000288368.5
|
PREX2
|
phosphatidylinositol-3,4,5-trisphosphate dependent Rac exchange factor 2 |
| chr2_-_196592671 | 0.02 |
ENST00000260983.8
ENST00000644030.1 |
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr17_+_38730301 | 0.02 |
ENST00000613478.2
|
CISD3
|
CDGSH iron sulfur domain 3 |
| chrX_+_10156960 | 0.02 |
ENST00000380833.9
|
CLCN4
|
chloride voltage-gated channel 4 |
| chr1_-_151459471 | 0.02 |
ENST00000271715.7
|
POGZ
|
pogo transposable element derived with ZNF domain |
| chr7_-_13986439 | 0.02 |
ENST00000443608.5
ENST00000438956.5 |
ETV1
|
ETS variant transcription factor 1 |
| chr7_+_143132069 | 0.02 |
ENST00000291009.4
|
PIP
|
prolactin induced protein |
| chr16_-_1963100 | 0.02 |
ENST00000526586.6
|
RPS2
|
ribosomal protein S2 |
| chr11_-_58844695 | 0.02 |
ENST00000287275.6
|
GLYATL2
|
glycine-N-acyltransferase like 2 |
| chr11_-_35419098 | 0.02 |
ENST00000606205.6
ENST00000645303.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr5_+_161850597 | 0.02 |
ENST00000634335.1
ENST00000635880.1 |
GABRA1
|
gamma-aminobutyric acid type A receptor subunit alpha1 |
| chr19_-_52171643 | 0.02 |
ENST00000597065.1
|
ZNF836
|
zinc finger protein 836 |
| chr3_+_41194741 | 0.02 |
ENST00000643541.1
ENST00000426215.5 ENST00000645210.1 ENST00000646381.1 ENST00000405570.6 ENST00000642248.1 ENST00000433400.6 |
CTNNB1
|
catenin beta 1 |
| chr4_+_168497066 | 0.02 |
ENST00000261509.10
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr6_+_29100609 | 0.02 |
ENST00000377171.3
|
OR2J1
|
olfactory receptor family 2 subfamily J member 1 |
| chr1_+_196977550 | 0.02 |
ENST00000256785.5
|
CFHR5
|
complement factor H related 5 |
| chr5_+_141421064 | 0.02 |
ENST00000518882.2
|
PCDHGA11
|
protocadherin gamma subfamily A, 11 |
| chr1_+_153217578 | 0.02 |
ENST00000368744.4
|
PRR9
|
proline rich 9 |
| chr9_-_114657791 | 0.02 |
ENST00000423632.3
|
TEX53
|
testis expressed 53 |
| chr4_+_155903688 | 0.02 |
ENST00000536354.3
|
TDO2
|
tryptophan 2,3-dioxygenase |
| chr4_-_156970903 | 0.02 |
ENST00000422544.2
|
PDGFC
|
platelet derived growth factor C |
| chr17_+_32991867 | 0.02 |
ENST00000394638.1
|
SPACA3
|
sperm acrosome associated 3 |
| chr14_-_23551729 | 0.02 |
ENST00000419474.5
|
ZFHX2
|
zinc finger homeobox 2 |
| chr4_+_168497044 | 0.02 |
ENST00000505667.6
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr19_-_48513161 | 0.02 |
ENST00000673139.1
|
LMTK3
|
lemur tyrosine kinase 3 |
| chr7_-_13986498 | 0.02 |
ENST00000420159.6
ENST00000399357.7 ENST00000403527.5 |
ETV1
|
ETS variant transcription factor 1 |
| chr9_-_92482499 | 0.02 |
ENST00000375544.7
|
ASPN
|
asporin |
| chr3_-_94028582 | 0.02 |
ENST00000315099.3
|
STX19
|
syntaxin 19 |
| chr19_+_45093140 | 0.02 |
ENST00000544069.2
ENST00000221462.9 |
PPP1R37
|
protein phosphatase 1 regulatory subunit 37 |
| chr17_-_43546323 | 0.02 |
ENST00000545954.5
ENST00000319349.10 |
ETV4
|
ETS variant transcription factor 4 |
| chr12_-_57818704 | 0.02 |
ENST00000549994.1
|
AVIL
|
advillin |
| chr9_-_74952904 | 0.02 |
ENST00000376854.6
|
C9orf40
|
chromosome 9 open reading frame 40 |
| chr19_+_37506931 | 0.01 |
ENST00000627814.3
ENST00000587143.5 |
ZNF793
|
zinc finger protein 793 |
| chr1_-_159714581 | 0.01 |
ENST00000255030.9
ENST00000437342.1 ENST00000368112.5 ENST00000368111.5 ENST00000368110.1 |
CRP
|
C-reactive protein |
| chr1_-_110607816 | 0.01 |
ENST00000485317.6
|
KCNA2
|
potassium voltage-gated channel subfamily A member 2 |
| chr1_+_248038172 | 0.01 |
ENST00000366479.4
|
OR2L2
|
olfactory receptor family 2 subfamily L member 2 |
| chr15_+_92900338 | 0.01 |
ENST00000625990.3
|
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr3_-_149221811 | 0.01 |
ENST00000455472.3
ENST00000264613.11 |
CP
|
ceruloplasmin |
| chr9_-_28670285 | 0.01 |
ENST00000379992.6
ENST00000308675.5 ENST00000613945.3 |
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr19_+_35449584 | 0.01 |
ENST00000246549.2
|
FFAR2
|
free fatty acid receptor 2 |
| chr15_+_38454103 | 0.01 |
ENST00000397609.6
ENST00000491535.5 |
FAM98B
|
family with sequence similarity 98 member B |
| chr14_+_21797272 | 0.01 |
ENST00000390430.2
|
TRAV8-1
|
T cell receptor alpha variable 8-1 |
| chr19_-_52171600 | 0.01 |
ENST00000682614.1
|
ZNF836
|
zinc finger protein 836 |
| chr8_-_81483226 | 0.01 |
ENST00000256104.5
|
FABP4
|
fatty acid binding protein 4 |
| chr2_-_55419565 | 0.01 |
ENST00000647341.1
ENST00000647401.1 ENST00000336838.10 ENST00000621814.4 ENST00000644033.1 ENST00000645477.1 ENST00000647517.1 |
CCDC88A
|
coiled-coil domain containing 88A |
| chr1_-_110607307 | 0.01 |
ENST00000639048.2
ENST00000675391.1 ENST00000639233.2 |
KCNA2
|
potassium voltage-gated channel subfamily A member 2 |
| chr16_+_25216943 | 0.01 |
ENST00000219660.6
|
AQP8
|
aquaporin 8 |
| chr1_-_110607425 | 0.01 |
ENST00000633222.1
|
KCNA2
|
potassium voltage-gated channel subfamily A member 2 |
| chr6_+_113857333 | 0.01 |
ENST00000612661.2
|
MARCKS
|
myristoylated alanine rich protein kinase C substrate |
| chr4_-_20984011 | 0.01 |
ENST00000382149.9
|
KCNIP4
|
potassium voltage-gated channel interacting protein 4 |
| chr3_+_63443306 | 0.01 |
ENST00000472899.5
ENST00000479198.5 ENST00000460711.5 ENST00000465156.1 |
SYNPR
|
synaptoporin |
| chr5_+_148394712 | 0.01 |
ENST00000513826.1
|
FBXO38
|
F-box protein 38 |
| chrX_+_101550537 | 0.01 |
ENST00000372829.8
|
ARMCX1
|
armadillo repeat containing X-linked 1 |
| chr4_+_70721953 | 0.01 |
ENST00000381006.8
ENST00000226328.8 |
RUFY3
|
RUN and FYVE domain containing 3 |
| chr17_+_81395469 | 0.01 |
ENST00000584436.7
|
BAHCC1
|
BAH domain and coiled-coil containing 1 |
| chr1_-_110607970 | 0.01 |
ENST00000638532.1
|
KCNA2
|
potassium voltage-gated channel subfamily A member 2 |
| chr16_-_30610342 | 0.01 |
ENST00000287461.8
|
ZNF689
|
zinc finger protein 689 |
| chr17_+_81395449 | 0.01 |
ENST00000675386.2
|
BAHCC1
|
BAH domain and coiled-coil containing 1 |
| chr3_-_71583592 | 0.01 |
ENST00000650156.1
ENST00000649596.1 |
FOXP1
|
forkhead box P1 |
| chr2_+_108588453 | 0.01 |
ENST00000393310.5
|
LIMS1
|
LIM zinc finger domain containing 1 |
| chr6_+_158312459 | 0.01 |
ENST00000367097.8
|
TULP4
|
TUB like protein 4 |
| chr9_+_72114595 | 0.01 |
ENST00000545168.5
|
GDA
|
guanine deaminase |
| chr18_+_32091849 | 0.01 |
ENST00000261593.8
ENST00000578914.1 |
RNF138
|
ring finger protein 138 |
| chr2_-_206086057 | 0.01 |
ENST00000403263.6
|
INO80D
|
INO80 complex subunit D |
| chr19_-_4535221 | 0.01 |
ENST00000381848.7
ENST00000586133.1 |
PLIN5
|
perilipin 5 |
| chr9_-_96655280 | 0.01 |
ENST00000446045.1
ENST00000375234.8 |
PRXL2C
|
peroxiredoxin like 2C |
| chr1_-_151459169 | 0.01 |
ENST00000368863.6
ENST00000409503.5 ENST00000491586.5 ENST00000533351.5 |
POGZ
|
pogo transposable element derived with ZNF domain |
| chr4_+_186266183 | 0.01 |
ENST00000403665.7
ENST00000492972.6 ENST00000264692.8 |
F11
|
coagulation factor XI |
| chr8_-_18684093 | 0.01 |
ENST00000428502.6
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chrX_+_46912276 | 0.01 |
ENST00000424392.5
ENST00000611250.4 |
JADE3
|
jade family PHD finger 3 |
| chr17_+_7558465 | 0.01 |
ENST00000349228.8
|
TNFSF13
|
TNF superfamily member 13 |
| chr5_-_136365476 | 0.01 |
ENST00000378459.7
ENST00000502753.4 ENST00000513104.6 ENST00000352189.8 |
TRPC7
|
transient receptor potential cation channel subfamily C member 7 |
| chr5_-_157345645 | 0.01 |
ENST00000312349.5
|
FNDC9
|
fibronectin type III domain containing 9 |
| chr2_+_67397297 | 0.01 |
ENST00000644028.1
ENST00000272342.6 |
ETAA1
|
ETAA1 activator of ATR kinase |
| chr8_+_76681208 | 0.01 |
ENST00000651372.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chrX_-_19970298 | 0.01 |
ENST00000379687.7
ENST00000379682.8 |
BCLAF3
|
BCLAF1 and THRAP3 family member 3 |
| chr7_-_93148345 | 0.01 |
ENST00000437805.5
ENST00000446959.5 ENST00000439952.5 ENST00000414791.5 ENST00000446033.1 ENST00000411955.5 ENST00000318238.9 |
SAMD9L
|
sterile alpha motif domain containing 9 like |
| chr8_-_18684033 | 0.01 |
ENST00000614430.3
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chrX_-_86047527 | 0.01 |
ENST00000615443.1
ENST00000357749.7 |
CHM
|
CHM Rab escort protein |
| chr1_-_162376841 | 0.01 |
ENST00000367935.10
|
SPATA46
|
spermatogenesis associated 46 |
| chr3_+_46242453 | 0.01 |
ENST00000452454.1
ENST00000395940.3 ENST00000457243.1 |
CCR3
|
C-C motif chemokine receptor 3 |
| chr4_+_157220691 | 0.01 |
ENST00000509417.5
ENST00000645636.1 ENST00000296526.12 ENST00000264426.14 |
GRIA2
|
glutamate ionotropic receptor AMPA type subunit 2 |
| chr14_+_61697622 | 0.01 |
ENST00000539097.2
|
HIF1A
|
hypoxia inducible factor 1 subunit alpha |
| chr9_-_41189310 | 0.01 |
ENST00000456520.5
ENST00000377391.8 ENST00000613716.4 ENST00000617933.1 |
CBWD6
|
COBW domain containing 6 |
| chrX_+_46912412 | 0.00 |
ENST00000614628.5
|
JADE3
|
jade family PHD finger 3 |
| chr2_-_162838728 | 0.00 |
ENST00000328032.8
ENST00000332142.10 |
KCNH7
|
potassium voltage-gated channel subfamily H member 7 |
| chr5_+_66144288 | 0.00 |
ENST00000334121.11
|
SREK1
|
splicing regulatory glutamic acid and lysine rich protein 1 |
| chr3_+_69936629 | 0.00 |
ENST00000394348.2
ENST00000531774.1 |
MITF
|
melanocyte inducing transcription factor |
| chr15_+_37934626 | 0.00 |
ENST00000559502.5
ENST00000558148.5 ENST00000319669.5 ENST00000558158.5 |
TMCO5A
|
transmembrane and coiled-coil domains 5A |
| chr3_-_11720728 | 0.00 |
ENST00000445411.5
ENST00000273038.7 |
VGLL4
|
vestigial like family member 4 |
| chr7_-_78771265 | 0.00 |
ENST00000630991.2
ENST00000629359.2 |
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr16_+_72054477 | 0.00 |
ENST00000355906.10
ENST00000570083.5 ENST00000228226.12 ENST00000398131.6 ENST00000569639.5 ENST00000564499.5 ENST00000357763.8 ENST00000613898.1 ENST00000562526.5 ENST00000565574.5 ENST00000568417.6 |
HP
|
haptoglobin |
| chr3_-_120093666 | 0.00 |
ENST00000316626.6
ENST00000650344.2 ENST00000678439.1 |
GSK3B
|
glycogen synthase kinase 3 beta |
| chr22_+_30080460 | 0.00 |
ENST00000336726.11
|
HORMAD2
|
HORMA domain containing 2 |
| chr2_+_33436304 | 0.00 |
ENST00000402538.7
|
RASGRP3
|
RAS guanyl releasing protein 3 |
| chr4_-_68670648 | 0.00 |
ENST00000338206.6
|
UGT2B15
|
UDP glucuronosyltransferase family 2 member B15 |
| chr10_+_69801892 | 0.00 |
ENST00000398978.8
ENST00000645393.2 ENST00000354547.7 ENST00000674121.1 ENST00000673842.1 ENST00000520267.5 |
COL13A1
|
collagen type XIII alpha 1 chain |
| chr1_-_31919368 | 0.00 |
ENST00000457805.6
ENST00000602725.5 ENST00000679970.1 |
PTP4A2
ENSG00000288678.1
|
protein tyrosine phosphatase 4A2 novel protein |
| chr4_+_157220654 | 0.00 |
ENST00000393815.6
|
GRIA2
|
glutamate ionotropic receptor AMPA type subunit 2 |
| chr13_-_35855758 | 0.00 |
ENST00000615680.4
|
DCLK1
|
doublecortin like kinase 1 |
| chr6_+_52420992 | 0.00 |
ENST00000636954.1
ENST00000636566.1 ENST00000638075.1 |
EFHC1
|
EF-hand domain containing 1 |
| chr2_-_55419276 | 0.00 |
ENST00000646796.1
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr17_-_69268812 | 0.00 |
ENST00000586811.1
|
ABCA5
|
ATP binding cassette subfamily A member 5 |
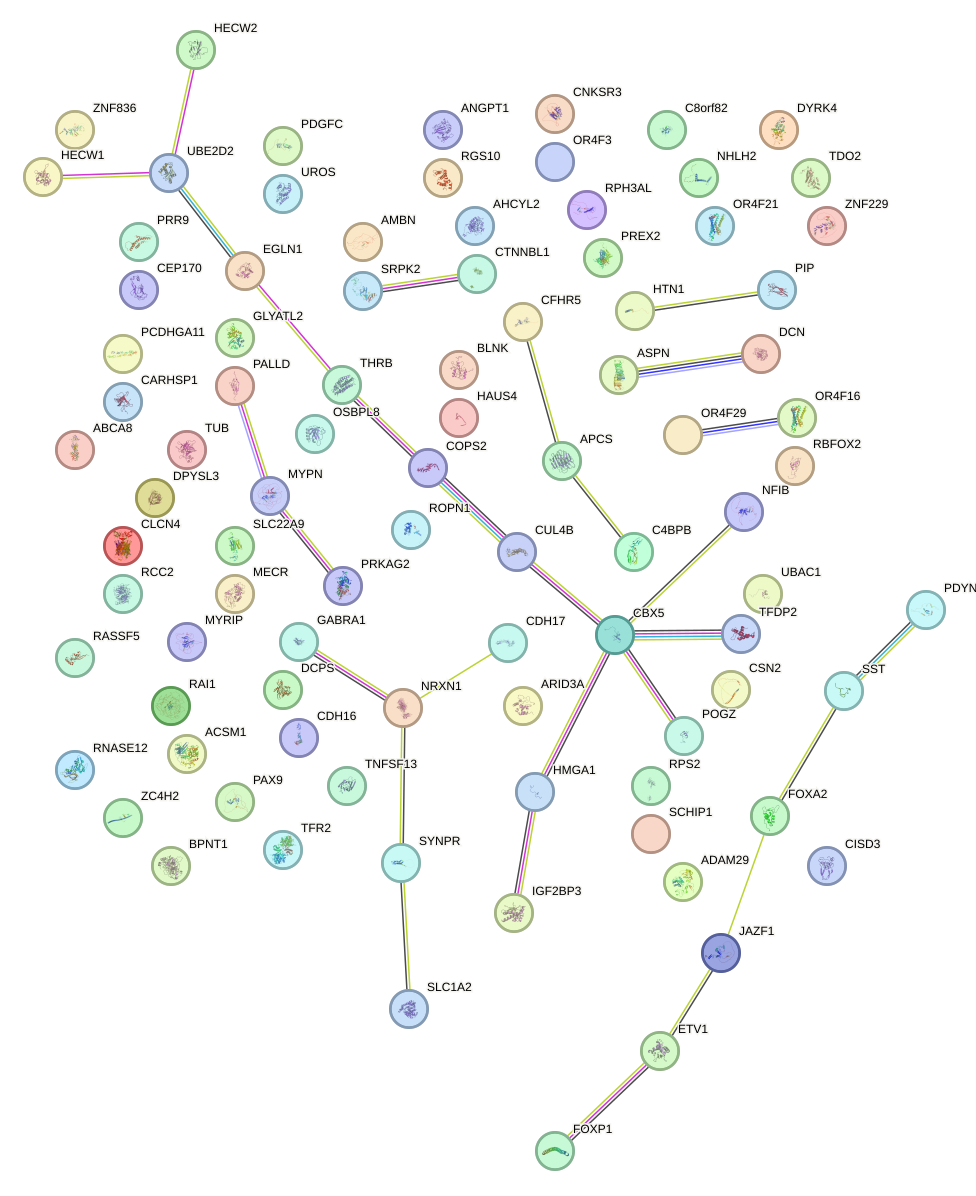
Network of associatons between targets according to the STRING database.
First level regulatory network of ONECUT2_ONECUT3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0035691 | macrophage migration inhibitory factor signaling pathway(GO:0035691) |
| 0.0 | 0.1 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.0 | 0.1 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.0 | 0.0 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.0 | 0.0 | GO:0036245 | cellular response to menadione(GO:0036245) |
| 0.0 | 0.0 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.0 | 0.0 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.0 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.1 | GO:1903027 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) regulation of opsonization(GO:1903027) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.1 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.0 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |