Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
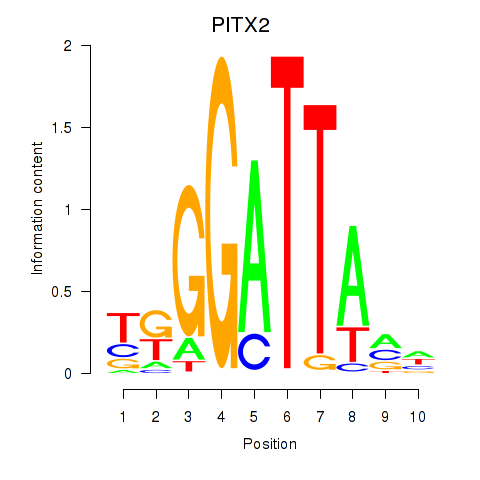
Results for PITX2
Z-value: 0.56
Transcription factors associated with PITX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PITX2
|
ENSG00000164093.17 | paired like homeodomain 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PITX2 | hg38_v1_chr4_-_110636963_110637042, hg38_v1_chr4_-_110623051_110623085, hg38_v1_chr4_-_110641920_110642123 | 0.56 | 1.5e-01 | Click! |
Activity profile of PITX2 motif
Sorted Z-values of PITX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_41518878 | 0.77 |
ENST00000254043.8
|
KRT15
|
keratin 15 |
| chr2_+_120013111 | 0.41 |
ENST00000331393.8
ENST00000443124.5 |
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr9_+_12693327 | 0.31 |
ENST00000388918.10
|
TYRP1
|
tyrosinase related protein 1 |
| chr17_-_4739866 | 0.21 |
ENST00000574412.6
ENST00000293778.12 |
CXCL16
|
C-X-C motif chemokine ligand 16 |
| chr6_-_138572523 | 0.20 |
ENST00000427025.6
|
NHSL1
|
NHS like 1 |
| chr6_+_130018565 | 0.14 |
ENST00000361794.7
ENST00000526087.5 ENST00000533560.5 |
L3MBTL3
|
L3MBTL histone methyl-lysine binding protein 3 |
| chr11_-_36598221 | 0.13 |
ENST00000311485.8
ENST00000527033.5 ENST00000532616.1 ENST00000618712.4 |
RAG2
|
recombination activating 2 |
| chr13_+_73058993 | 0.11 |
ENST00000377687.6
|
KLF5
|
Kruppel like factor 5 |
| chr3_-_191282383 | 0.11 |
ENST00000427544.6
|
UTS2B
|
urotensin 2B |
| chr7_+_30284574 | 0.09 |
ENST00000323037.5
|
ZNRF2
|
zinc and ring finger 2 |
| chr1_+_43300971 | 0.09 |
ENST00000372476.8
ENST00000538015.1 |
TIE1
|
tyrosine kinase with immunoglobulin like and EGF like domains 1 |
| chr21_-_33542841 | 0.09 |
ENST00000381831.7
ENST00000381839.7 |
GART
|
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
| chr6_+_31137646 | 0.08 |
ENST00000547221.1
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1 |
| chr13_-_35855627 | 0.08 |
ENST00000379893.5
|
DCLK1
|
doublecortin like kinase 1 |
| chr15_+_40929338 | 0.08 |
ENST00000249749.7
|
DLL4
|
delta like canonical Notch ligand 4 |
| chr12_+_69739370 | 0.07 |
ENST00000550536.5
ENST00000362025.9 |
RAB3IP
|
RAB3A interacting protein |
| chr11_-_5227063 | 0.07 |
ENST00000335295.4
ENST00000485743.1 ENST00000647020.1 |
HBB
|
hemoglobin subunit beta |
| chr6_-_53148822 | 0.06 |
ENST00000259803.8
|
GCM1
|
glial cells missing transcription factor 1 |
| chr10_-_60141004 | 0.05 |
ENST00000355288.6
|
ANK3
|
ankyrin 3 |
| chr16_-_67980483 | 0.05 |
ENST00000268793.6
ENST00000672962.1 |
DPEP3
|
dipeptidase 3 |
| chr15_+_41774539 | 0.05 |
ENST00000514566.5
|
MAPKBP1
|
mitogen-activated protein kinase binding protein 1 |
| chr12_-_54981838 | 0.04 |
ENST00000316577.12
|
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr5_+_38258373 | 0.04 |
ENST00000354891.7
|
EGFLAM
|
EGF like, fibronectin type III and laminin G domains |
| chr1_+_202010575 | 0.04 |
ENST00000367283.7
ENST00000367284.10 |
ELF3
|
E74 like ETS transcription factor 3 |
| chr7_-_122699108 | 0.04 |
ENST00000340112.3
|
RNF133
|
ring finger protein 133 |
| chr4_-_101346842 | 0.04 |
ENST00000507176.5
|
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha |
| chr13_-_35855758 | 0.03 |
ENST00000615680.4
|
DCLK1
|
doublecortin like kinase 1 |
| chr9_-_128724088 | 0.03 |
ENST00000406904.2
ENST00000452105.5 ENST00000372667.9 ENST00000372663.9 |
ZDHHC12
|
zinc finger DHHC-type palmitoyltransferase 12 |
| chr6_+_35342614 | 0.03 |
ENST00000337400.6
ENST00000311565.4 |
PPARD
|
peroxisome proliferator activated receptor delta |
| chr17_+_4740042 | 0.03 |
ENST00000592813.5
|
ZMYND15
|
zinc finger MYND-type containing 15 |
| chr4_-_101347492 | 0.03 |
ENST00000394854.8
|
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha |
| chr17_+_4740005 | 0.03 |
ENST00000269289.10
|
ZMYND15
|
zinc finger MYND-type containing 15 |
| chr4_-_101347327 | 0.03 |
ENST00000394853.8
|
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha |
| chr13_-_94479671 | 0.03 |
ENST00000377028.10
ENST00000446125.1 |
DCT
|
dopachrome tautomerase |
| chrX_+_15749848 | 0.03 |
ENST00000479740.5
ENST00000454127.2 |
CA5B
|
carbonic anhydrase 5B |
| chr1_+_2476315 | 0.03 |
ENST00000419816.6
|
PLCH2
|
phospholipase C eta 2 |
| chr1_+_91952162 | 0.03 |
ENST00000402388.1
ENST00000680541.1 |
BRDT
|
bromodomain testis associated |
| chr1_+_2476284 | 0.03 |
ENST00000378486.8
|
PLCH2
|
phospholipase C eta 2 |
| chr4_-_101347471 | 0.03 |
ENST00000323055.10
ENST00000512215.5 |
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha |
| chr22_-_29838227 | 0.02 |
ENST00000307790.8
ENST00000397771.6 ENST00000542393.5 |
ASCC2
|
activating signal cointegrator 1 complex subunit 2 |
| chr11_-_133532493 | 0.02 |
ENST00000524381.6
|
OPCML
|
opioid binding protein/cell adhesion molecule like |
| chr12_-_118359639 | 0.02 |
ENST00000541786.5
ENST00000419821.6 ENST00000541878.5 |
TAOK3
|
TAO kinase 3 |
| chr12_+_6840800 | 0.02 |
ENST00000541978.5
ENST00000229264.8 ENST00000435982.6 |
GNB3
|
G protein subunit beta 3 |
| chrM_+_9207 | 0.02 |
ENST00000362079.2
|
MT-CO3
|
mitochondrially encoded cytochrome c oxidase III |
| chr14_+_75522531 | 0.02 |
ENST00000555504.1
|
BATF
|
basic leucine zipper ATF-like transcription factor |
| chr2_+_167248638 | 0.02 |
ENST00000295237.10
|
XIRP2
|
xin actin binding repeat containing 2 |
| chr18_-_49491586 | 0.02 |
ENST00000584895.5
ENST00000580210.5 ENST00000579408.5 |
RPL17-C18orf32
RPL17
|
RPL17-C18orf32 readthrough ribosomal protein L17 |
| chr9_-_14300231 | 0.01 |
ENST00000636735.1
|
NFIB
|
nuclear factor I B |
| chr12_-_52796110 | 0.01 |
ENST00000417996.2
|
KRT3
|
keratin 3 |
| chr16_+_28711417 | 0.01 |
ENST00000395587.5
ENST00000569690.5 ENST00000331666.11 ENST00000564243.5 ENST00000566866.5 |
EIF3C
|
eukaryotic translation initiation factor 3 subunit C |
| chr2_-_65432591 | 0.01 |
ENST00000356388.9
|
SPRED2
|
sprouty related EVH1 domain containing 2 |
| chrX_-_6228835 | 0.01 |
ENST00000381095.8
|
NLGN4X
|
neuroligin 4 X-linked |
| chr16_+_57976435 | 0.01 |
ENST00000290871.10
ENST00000441824.4 |
TEPP
|
testis, prostate and placenta expressed |
| chr15_-_89221558 | 0.01 |
ENST00000268125.10
|
RLBP1
|
retinaldehyde binding protein 1 |
| chr8_-_132111159 | 0.01 |
ENST00000673615.1
ENST00000434736.6 |
HHLA1
|
HERV-H LTR-associating 1 |
| chr7_+_155458129 | 0.01 |
ENST00000297375.4
|
EN2
|
engrailed homeobox 2 |
| chr6_+_35342535 | 0.01 |
ENST00000360694.8
ENST00000418635.6 ENST00000448077.6 |
PPARD
|
peroxisome proliferator activated receptor delta |
| chr22_-_38700655 | 0.01 |
ENST00000216039.9
|
JOSD1
|
Josephin domain containing 1 |
| chr10_-_73433550 | 0.00 |
ENST00000299432.7
|
MSS51
|
MSS51 mitochondrial translational activator |
| chr19_+_47821907 | 0.00 |
ENST00000539067.5
ENST00000221996.12 ENST00000613299.1 |
CRX
|
cone-rod homeobox |
| chr8_+_48008409 | 0.00 |
ENST00000523432.5
ENST00000521346.5 ENST00000523111.7 ENST00000517630.5 |
UBE2V2
|
ubiquitin conjugating enzyme E2 V2 |
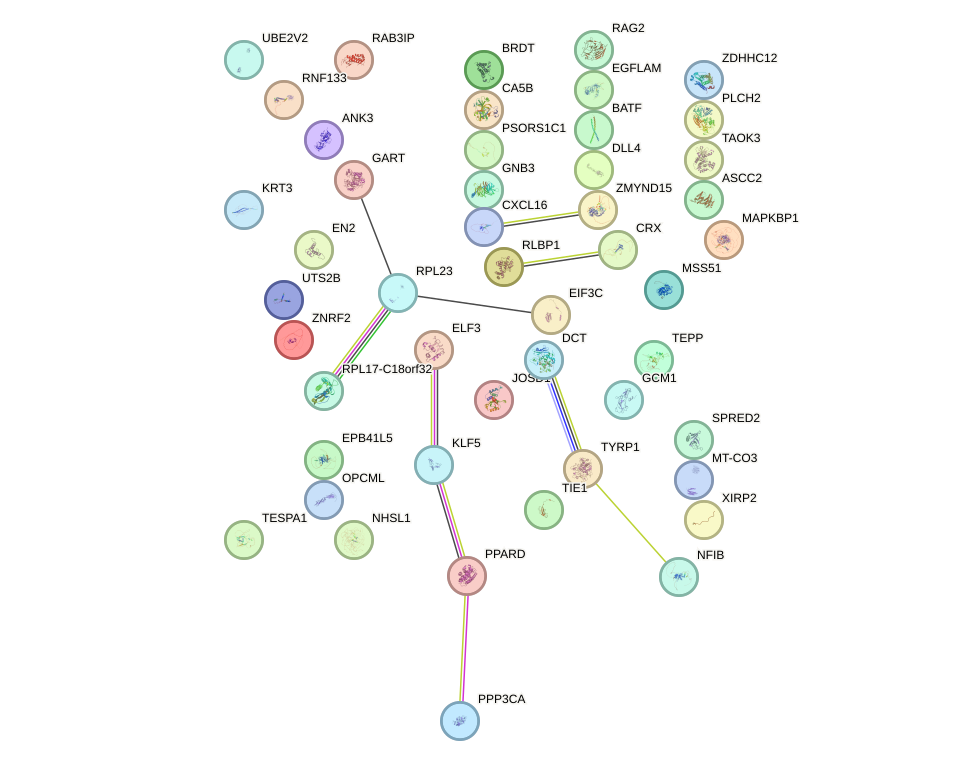
Network of associatons between targets according to the STRING database.
First level regulatory network of PITX2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0048319 | mesodermal cell migration(GO:0008078) axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.1 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.1 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.0 | 0.3 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.1 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.0 | 0.2 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.1 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.0 | 0.1 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.0 | 0.1 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.0 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.1 | GO:0043249 | erythrocyte maturation(GO:0043249) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.3 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |