Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
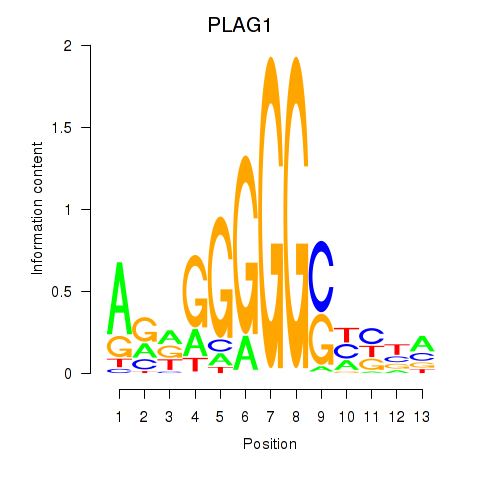
Results for PLAG1
Z-value: 0.33
Transcription factors associated with PLAG1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PLAG1
|
ENSG00000181690.8 | PLAG1 zinc finger |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PLAG1 | hg38_v1_chr8_-_56211257_56211309 | 0.67 | 6.8e-02 | Click! |
Activity profile of PLAG1 motif
Sorted Z-values of PLAG1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_163068775 | 0.35 |
ENST00000421743.6
|
RGS4
|
regulator of G protein signaling 4 |
| chr11_+_46277648 | 0.32 |
ENST00000621158.5
|
CREB3L1
|
cAMP responsive element binding protein 3 like 1 |
| chr12_-_108339300 | 0.23 |
ENST00000550402.6
ENST00000552995.5 ENST00000312143.11 |
CMKLR1
|
chemerin chemokine-like receptor 1 |
| chr12_+_78864768 | 0.21 |
ENST00000261205.9
ENST00000457153.6 |
SYT1
|
synaptotagmin 1 |
| chr4_+_54229261 | 0.20 |
ENST00000508170.5
ENST00000512143.1 ENST00000257290.10 |
PDGFRA
|
platelet derived growth factor receptor alpha |
| chr5_-_138033021 | 0.19 |
ENST00000033079.7
|
FAM13B
|
family with sequence similarity 13 member B |
| chr14_-_74612226 | 0.17 |
ENST00000261978.9
|
LTBP2
|
latent transforming growth factor beta binding protein 2 |
| chrX_+_52184874 | 0.17 |
ENST00000599522.7
ENST00000471932.6 |
MAGED4
|
MAGE family member D4 |
| chr5_-_55534955 | 0.17 |
ENST00000307259.9
ENST00000264775.9 |
PLPP1
|
phospholipid phosphatase 1 |
| chrX_+_150361559 | 0.15 |
ENST00000262858.8
|
MAMLD1
|
mastermind like domain containing 1 |
| chr11_-_57514876 | 0.14 |
ENST00000528450.5
|
SLC43A1
|
solute carrier family 43 member 1 |
| chr4_+_61200318 | 0.14 |
ENST00000683033.1
|
ADGRL3
|
adhesion G protein-coupled receptor L3 |
| chr3_-_73624840 | 0.13 |
ENST00000308537.4
ENST00000263666.9 |
PDZRN3
|
PDZ domain containing ring finger 3 |
| chrX_-_143636094 | 0.12 |
ENST00000356928.2
|
SLITRK4
|
SLIT and NTRK like family member 4 |
| chr12_-_48004496 | 0.12 |
ENST00000337299.7
|
COL2A1
|
collagen type II alpha 1 chain |
| chrX_+_52184904 | 0.12 |
ENST00000375626.7
ENST00000467526.1 |
MAGED4
|
MAGE family member D4 |
| chr9_+_136980211 | 0.12 |
ENST00000444903.2
|
PTGDS
|
prostaglandin D2 synthase |
| chr11_+_118607579 | 0.12 |
ENST00000530708.4
|
PHLDB1
|
pleckstrin homology like domain family B member 1 |
| chr17_+_83079595 | 0.11 |
ENST00000320095.12
|
METRNL
|
meteorin like, glial cell differentiation regulator |
| chr3_+_42654470 | 0.11 |
ENST00000680014.1
|
ZBTB47
|
zinc finger and BTB domain containing 47 |
| chr3_-_15332526 | 0.11 |
ENST00000383791.8
|
SH3BP5
|
SH3 domain binding protein 5 |
| chr15_+_22094522 | 0.11 |
ENST00000328795.5
|
OR4N4
|
olfactory receptor family 4 subfamily N member 4 |
| chr16_-_54286763 | 0.10 |
ENST00000329734.4
|
IRX3
|
iroquois homeobox 3 |
| chr16_+_31472130 | 0.10 |
ENST00000394863.8
ENST00000565360.5 ENST00000361773.7 |
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1 |
| chr19_+_39413528 | 0.10 |
ENST00000438123.5
ENST00000409797.6 ENST00000451354.6 |
PLEKHG2
|
pleckstrin homology and RhoGEF domain containing G2 |
| chr10_+_110007964 | 0.10 |
ENST00000277900.12
ENST00000356080.9 |
ADD3
|
adducin 3 |
| chr20_+_38724478 | 0.09 |
ENST00000217420.2
|
SLC32A1
|
solute carrier family 32 member 1 |
| chrX_-_16869840 | 0.09 |
ENST00000380084.8
|
RBBP7
|
RB binding protein 7, chromatin remodeling factor |
| chr5_-_59893718 | 0.09 |
ENST00000340635.11
|
PDE4D
|
phosphodiesterase 4D |
| chr4_-_8127650 | 0.09 |
ENST00000545242.6
ENST00000676532.1 |
ABLIM2
|
actin binding LIM protein family member 2 |
| chr1_-_68232539 | 0.09 |
ENST00000370976.7
ENST00000354777.6 |
WLS
|
Wnt ligand secretion mediator |
| chr19_+_3359563 | 0.08 |
ENST00000589123.5
ENST00000395111.7 ENST00000586919.5 |
NFIC
|
nuclear factor I C |
| chr3_+_69763726 | 0.08 |
ENST00000448226.9
|
MITF
|
melanocyte inducing transcription factor |
| chr19_-_51082883 | 0.08 |
ENST00000650543.2
|
KLK14
|
kallikrein related peptidase 14 |
| chr7_-_100573865 | 0.08 |
ENST00000622764.3
|
SAP25
|
Sin3A associated protein 25 |
| chr2_-_127642131 | 0.07 |
ENST00000426981.5
|
LIMS2
|
LIM zinc finger domain containing 2 |
| chr7_+_101085464 | 0.07 |
ENST00000306085.11
ENST00000412507.1 |
TRIM56
|
tripartite motif containing 56 |
| chr6_+_26500296 | 0.07 |
ENST00000684113.1
|
BTN1A1
|
butyrophilin subfamily 1 member A1 |
| chr16_-_29899245 | 0.07 |
ENST00000537485.5
|
SEZ6L2
|
seizure related 6 homolog like 2 |
| chr12_-_57738740 | 0.07 |
ENST00000547588.6
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr1_-_31704001 | 0.07 |
ENST00000373672.8
|
COL16A1
|
collagen type XVI alpha 1 chain |
| chr1_+_6448448 | 0.07 |
ENST00000475228.6
|
ESPN
|
espin |
| chr5_-_177509814 | 0.07 |
ENST00000510898.7
ENST00000502885.5 ENST00000506493.5 |
DOK3
|
docking protein 3 |
| chr10_+_92691897 | 0.07 |
ENST00000492654.3
|
HHEX
|
hematopoietically expressed homeobox |
| chr12_-_12266769 | 0.07 |
ENST00000543091.1
|
LRP6
|
LDL receptor related protein 6 |
| chr16_-_29899043 | 0.06 |
ENST00000346932.9
ENST00000350527.7 ENST00000568380.1 |
SEZ6L2
|
seizure related 6 homolog like 2 |
| chr17_-_76141240 | 0.06 |
ENST00000322957.7
|
FOXJ1
|
forkhead box J1 |
| chr8_-_102655707 | 0.06 |
ENST00000285407.11
|
KLF10
|
Kruppel like factor 10 |
| chr9_-_120714457 | 0.06 |
ENST00000373930.4
|
MEGF9
|
multiple EGF like domains 9 |
| chr17_+_75110021 | 0.06 |
ENST00000584947.1
|
ARMC7
|
armadillo repeat containing 7 |
| chr16_+_285680 | 0.06 |
ENST00000435833.1
|
PDIA2
|
protein disulfide isomerase family A member 2 |
| chr10_+_101131284 | 0.06 |
ENST00000370196.11
ENST00000467928.2 |
TLX1
|
T cell leukemia homeobox 1 |
| chr19_-_13906062 | 0.06 |
ENST00000591586.5
ENST00000346736.6 |
BRME1
|
break repair meiotic recombinase recruitment factor 1 |
| chr5_+_161848112 | 0.06 |
ENST00000393943.10
|
GABRA1
|
gamma-aminobutyric acid type A receptor subunit alpha1 |
| chr7_-_72969466 | 0.06 |
ENST00000285805.3
|
TRIM74
|
tripartite motif containing 74 |
| chr11_+_54706832 | 0.06 |
ENST00000319760.8
|
OR4A5
|
olfactory receptor family 4 subfamily A member 5 |
| chr16_-_29899532 | 0.06 |
ENST00000308713.9
ENST00000617533.5 |
SEZ6L2
|
seizure related 6 homolog like 2 |
| chr8_-_143976722 | 0.06 |
ENST00000436759.6
ENST00000527303.2 |
PLEC
|
plectin |
| chr7_+_75395629 | 0.06 |
ENST00000323819.7
ENST00000430211.5 |
TRIM73
|
tripartite motif containing 73 |
| chr11_-_73598067 | 0.06 |
ENST00000450446.6
ENST00000356467.5 |
FAM168A
|
family with sequence similarity 168 member A |
| chr1_+_150257247 | 0.06 |
ENST00000647854.1
|
CA14
|
carbonic anhydrase 14 |
| chr1_-_19980416 | 0.06 |
ENST00000375111.7
|
PLA2G2A
|
phospholipase A2 group IIA |
| chr11_-_64744317 | 0.06 |
ENST00000419843.1
ENST00000394430.5 |
RASGRP2
|
RAS guanyl releasing protein 2 |
| chr6_+_31715339 | 0.06 |
ENST00000375824.1
ENST00000375825.7 |
LY6G6D
|
lymphocyte antigen 6 family member G6D |
| chr12_-_112382363 | 0.06 |
ENST00000682272.1
ENST00000377560.9 |
HECTD4
|
HECT domain E3 ubiquitin protein ligase 4 |
| chr12_+_6452024 | 0.05 |
ENST00000266556.8
ENST00000544021.5 |
TAPBPL
|
TAP binding protein like |
| chr14_-_50830479 | 0.05 |
ENST00000382043.8
|
NIN
|
ninein |
| chr22_-_50307598 | 0.05 |
ENST00000425954.1
ENST00000449103.5 ENST00000359337.9 |
PLXNB2
|
plexin B2 |
| chr5_+_161848314 | 0.05 |
ENST00000437025.6
|
GABRA1
|
gamma-aminobutyric acid type A receptor subunit alpha1 |
| chrX_+_23664251 | 0.05 |
ENST00000379349.5
|
PRDX4
|
peroxiredoxin 4 |
| chr1_-_68232514 | 0.05 |
ENST00000262348.9
ENST00000370973.2 ENST00000370971.1 |
WLS
|
Wnt ligand secretion mediator |
| chr2_-_74391837 | 0.05 |
ENST00000417090.1
ENST00000409868.5 ENST00000680606.1 |
DCTN1
|
dynactin subunit 1 |
| chr16_-_30096170 | 0.05 |
ENST00000566134.5
ENST00000565110.5 ENST00000398841.6 ENST00000398838.8 |
YPEL3
|
yippee like 3 |
| chr7_-_73328082 | 0.05 |
ENST00000333149.7
|
TRIM50
|
tripartite motif containing 50 |
| chrX_+_111096136 | 0.05 |
ENST00000372007.10
|
PAK3
|
p21 (RAC1) activated kinase 3 |
| chr1_-_160070150 | 0.05 |
ENST00000644903.1
|
KCNJ10
|
potassium inwardly rectifying channel subfamily J member 10 |
| chr15_-_43618240 | 0.05 |
ENST00000432436.1
|
STRC
|
stereocilin |
| chr2_-_85561481 | 0.05 |
ENST00000233838.9
|
GGCX
|
gamma-glutamyl carboxylase |
| chr11_-_73598183 | 0.05 |
ENST00000064778.8
|
FAM168A
|
family with sequence similarity 168 member A |
| chr14_+_79279339 | 0.05 |
ENST00000557594.5
|
NRXN3
|
neurexin 3 |
| chr14_-_57866075 | 0.05 |
ENST00000556826.6
|
SLC35F4
|
solute carrier family 35 member F4 |
| chr1_+_151166001 | 0.05 |
ENST00000368902.1
ENST00000368905.9 |
SCNM1
|
sodium channel modifier 1 |
| chrX_+_111096211 | 0.05 |
ENST00000372010.5
ENST00000519681.5 |
PAK3
|
p21 (RAC1) activated kinase 3 |
| chr20_+_58891981 | 0.04 |
ENST00000488652.6
ENST00000476935.6 ENST00000492907.6 ENST00000603546.2 |
GNAS
|
GNAS complex locus |
| chr22_-_30246739 | 0.04 |
ENST00000403987.3
ENST00000249075.4 |
LIF
|
LIF interleukin 6 family cytokine |
| chr17_-_4704040 | 0.04 |
ENST00000570571.5
ENST00000575101.1 ENST00000574876.5 ENST00000572293.7 |
PELP1
|
proline, glutamate and leucine rich protein 1 |
| chr7_+_75395063 | 0.04 |
ENST00000450434.5
|
TRIM73
|
tripartite motif containing 73 |
| chr6_+_32844789 | 0.04 |
ENST00000414474.5
|
PSMB9
|
proteasome 20S subunit beta 9 |
| chr1_+_145964675 | 0.04 |
ENST00000369314.2
ENST00000369313.7 |
POLR3GL
|
RNA polymerase III subunit GL |
| chr14_+_79279681 | 0.04 |
ENST00000679122.1
|
NRXN3
|
neurexin 3 |
| chr10_+_17229267 | 0.04 |
ENST00000224237.9
|
VIM
|
vimentin |
| chr14_+_79279403 | 0.04 |
ENST00000281127.11
|
NRXN3
|
neurexin 3 |
| chr2_-_29921580 | 0.04 |
ENST00000389048.8
|
ALK
|
ALK receptor tyrosine kinase |
| chr22_-_30387078 | 0.04 |
ENST00000215798.10
|
RNF215
|
ring finger protein 215 |
| chr1_-_160070102 | 0.04 |
ENST00000638728.1
ENST00000637644.1 |
KCNJ10
|
potassium inwardly rectifying channel subfamily J member 10 |
| chr9_-_137032081 | 0.04 |
ENST00000314412.7
|
FUT7
|
fucosyltransferase 7 |
| chr6_-_35497042 | 0.04 |
ENST00000639578.3
ENST00000338863.13 |
TEAD3
|
TEA domain transcription factor 3 |
| chr4_-_113761724 | 0.04 |
ENST00000511664.6
|
CAMK2D
|
calcium/calmodulin dependent protein kinase II delta |
| chr4_+_41990496 | 0.04 |
ENST00000264451.12
|
SLC30A9
|
solute carrier family 30 member 9 |
| chr14_-_25050111 | 0.04 |
ENST00000323944.9
|
STXBP6
|
syntaxin binding protein 6 |
| chr6_+_31575557 | 0.04 |
ENST00000449264.3
|
TNF
|
tumor necrosis factor |
| chr9_+_136327526 | 0.04 |
ENST00000440944.6
|
GPSM1
|
G protein signaling modulator 1 |
| chr16_-_57536543 | 0.04 |
ENST00000258214.3
|
CCDC102A
|
coiled-coil domain containing 102A |
| chr4_-_113761441 | 0.04 |
ENST00000394524.7
|
CAMK2D
|
calcium/calmodulin dependent protein kinase II delta |
| chr19_+_55385928 | 0.04 |
ENST00000431533.6
ENST00000428193.6 ENST00000558815.5 ENST00000344063.7 ENST00000560583.5 ENST00000560055.5 ENST00000559463.5 |
RPL28
|
ribosomal protein L28 |
| chr19_+_38433676 | 0.04 |
ENST00000359596.8
ENST00000355481.8 |
RYR1
|
ryanodine receptor 1 |
| chr15_+_40894410 | 0.03 |
ENST00000220509.10
ENST00000558474.1 |
VPS18
|
VPS18 core subunit of CORVET and HOPS complexes |
| chr14_+_79279906 | 0.03 |
ENST00000428277.6
|
NRXN3
|
neurexin 3 |
| chr17_+_75109939 | 0.03 |
ENST00000581078.1
ENST00000245543.6 ENST00000582136.5 |
ARMC7
|
armadillo repeat containing 7 |
| chr3_+_135965718 | 0.03 |
ENST00000264977.8
ENST00000490467.5 |
PPP2R3A
|
protein phosphatase 2 regulatory subunit B''alpha |
| chr14_+_104581141 | 0.03 |
ENST00000410013.1
|
C14orf180
|
chromosome 14 open reading frame 180 |
| chr10_+_47322450 | 0.03 |
ENST00000581492.3
|
GDF2
|
growth differentiation factor 2 |
| chr10_+_134465 | 0.03 |
ENST00000439456.5
ENST00000397962.8 ENST00000309776.8 ENST00000397959.7 |
ZMYND11
|
zinc finger MYND-type containing 11 |
| chr5_+_110738983 | 0.03 |
ENST00000355943.8
ENST00000447245.6 |
SLC25A46
|
solute carrier family 25 member 46 |
| chr2_-_178072751 | 0.03 |
ENST00000286063.11
|
PDE11A
|
phosphodiesterase 11A |
| chr9_-_137114678 | 0.03 |
ENST00000497375.1
ENST00000371579.7 |
DPP7
|
dipeptidyl peptidase 7 |
| chr2_-_55419276 | 0.03 |
ENST00000646796.1
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr1_-_115338231 | 0.03 |
ENST00000369512.3
ENST00000680116.1 ENST00000681124.1 ENST00000675637.2 ENST00000676038.2 |
NGF
|
nerve growth factor |
| chr6_-_166308385 | 0.03 |
ENST00000322583.5
|
PRR18
|
proline rich 18 |
| chr7_+_6637311 | 0.03 |
ENST00000382252.6
|
ZNF316
|
zinc finger protein 316 |
| chrX_-_72307148 | 0.03 |
ENST00000453707.6
ENST00000373619.7 |
CITED1
|
Cbp/p300 interacting transactivator with Glu/Asp rich carboxy-terminal domain 1 |
| chr19_-_4535221 | 0.03 |
ENST00000381848.7
ENST00000586133.1 |
PLIN5
|
perilipin 5 |
| chr4_-_113761927 | 0.03 |
ENST00000296402.9
|
CAMK2D
|
calcium/calmodulin dependent protein kinase II delta |
| chr17_+_20579724 | 0.03 |
ENST00000661883.1
ENST00000399044.1 |
CDRT15L2
|
CMT1A duplicated region transcript 15 like 2 |
| chr16_+_14833713 | 0.03 |
ENST00000287667.12
ENST00000620755.4 ENST00000610363.4 |
NOMO1
|
NODAL modulator 1 |
| chr4_-_16226460 | 0.03 |
ENST00000405303.7
|
TAPT1
|
transmembrane anterior posterior transformation 1 |
| chr1_+_202462730 | 0.03 |
ENST00000290419.9
ENST00000491336.5 |
PPP1R12B
|
protein phosphatase 1 regulatory subunit 12B |
| chr5_+_134846065 | 0.03 |
ENST00000504727.1
ENST00000435259.2 ENST00000508791.1 |
C5orf24
|
chromosome 5 open reading frame 24 |
| chr8_-_130443581 | 0.03 |
ENST00000357668.2
ENST00000518721.6 |
ASAP1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
| chr2_-_74392025 | 0.03 |
ENST00000440727.1
ENST00000409240.5 |
DCTN1
|
dynactin subunit 1 |
| chr11_-_95231046 | 0.02 |
ENST00000416495.6
ENST00000536441.7 |
SESN3
|
sestrin 3 |
| chr7_-_156892987 | 0.02 |
ENST00000415428.5
|
LMBR1
|
limb development membrane protein 1 |
| chr11_+_57338344 | 0.02 |
ENST00000263314.3
ENST00000616487.4 |
P2RX3
|
purinergic receptor P2X 3 |
| chr1_-_44017296 | 0.02 |
ENST00000357730.6
ENST00000360584.6 ENST00000528803.1 |
SLC6A9
|
solute carrier family 6 member 9 |
| chr7_+_73328177 | 0.02 |
ENST00000442793.5
ENST00000413573.6 |
FKBP6
|
FKBP prolyl isomerase family member 6 (inactive) |
| chr14_-_25049889 | 0.02 |
ENST00000419632.6
ENST00000396700.5 |
STXBP6
|
syntaxin binding protein 6 |
| chr19_-_35135180 | 0.02 |
ENST00000392225.7
|
LGI4
|
leucine rich repeat LGI family member 4 |
| chr12_+_56152579 | 0.02 |
ENST00000551834.5
ENST00000552568.5 |
MYL6B
|
myosin light chain 6B |
| chr16_+_2471284 | 0.02 |
ENST00000293973.2
|
NTN3
|
netrin 3 |
| chr2_-_85561513 | 0.02 |
ENST00000430215.7
|
GGCX
|
gamma-glutamyl carboxylase |
| chr4_+_36281591 | 0.02 |
ENST00000639862.2
ENST00000357504.7 |
DTHD1
|
death domain containing 1 |
| chr7_+_73328152 | 0.02 |
ENST00000252037.5
ENST00000431982.6 |
FKBP6
|
FKBP prolyl isomerase family member 6 (inactive) |
| chr7_-_158587710 | 0.02 |
ENST00000389416.8
|
PTPRN2
|
protein tyrosine phosphatase receptor type N2 |
| chr17_+_4951080 | 0.02 |
ENST00000521811.5
ENST00000323997.10 ENST00000522249.5 ENST00000519584.5 ENST00000519602.6 |
ENO3
|
enolase 3 |
| chr12_-_27780236 | 0.02 |
ENST00000381273.4
|
MANSC4
|
MANSC domain containing 4 |
| chr19_+_55386338 | 0.02 |
ENST00000558131.1
ENST00000558752.1 |
RPL28
|
ribosomal protein L28 |
| chr16_-_18801424 | 0.02 |
ENST00000546206.6
ENST00000562819.5 ENST00000304414.12 ENST00000562234.2 ENST00000567078.2 |
ARL6IP1
ENSG00000260342.2
|
ADP ribosylation factor like GTPase 6 interacting protein 1 novel protein |
| chr12_+_57611420 | 0.02 |
ENST00000286494.9
|
ARHGEF25
|
Rho guanine nucleotide exchange factor 25 |
| chr21_+_44646414 | 0.02 |
ENST00000334670.9
|
KRTAP10-11
|
keratin associated protein 10-11 |
| chr12_+_51888217 | 0.02 |
ENST00000340970.8
|
ANKRD33
|
ankyrin repeat domain 33 |
| chr13_+_53028806 | 0.02 |
ENST00000219022.3
|
OLFM4
|
olfactomedin 4 |
| chr19_-_461007 | 0.02 |
ENST00000264554.11
|
SHC2
|
SHC adaptor protein 2 |
| chr2_-_152098670 | 0.02 |
ENST00000636129.1
ENST00000636785.1 ENST00000636496.1 |
CACNB4
|
calcium voltage-gated channel auxiliary subunit beta 4 |
| chr19_-_38229714 | 0.02 |
ENST00000416611.5
|
DPF1
|
double PHD fingers 1 |
| chr1_+_26993684 | 0.02 |
ENST00000522111.3
|
TRNP1
|
TMF1 regulated nuclear protein 1 |
| chr11_+_64285839 | 0.02 |
ENST00000438980.7
ENST00000540370.1 |
GPR137
|
G protein-coupled receptor 137 |
| chr19_-_35135011 | 0.02 |
ENST00000310123.8
|
LGI4
|
leucine rich repeat LGI family member 4 |
| chr3_-_116444983 | 0.02 |
ENST00000333617.8
|
LSAMP
|
limbic system associated membrane protein |
| chr8_-_23404076 | 0.02 |
ENST00000524168.1
ENST00000389131.8 ENST00000523833.2 ENST00000519243.1 |
LOXL2
|
lysyl oxidase like 2 |
| chrX_-_48919015 | 0.02 |
ENST00000376509.4
|
PIM2
|
Pim-2 proto-oncogene, serine/threonine kinase |
| chr9_+_129081098 | 0.02 |
ENST00000406974.7
ENST00000372546.9 |
DOLPP1
|
dolichyldiphosphatase 1 |
| chr11_+_66843413 | 0.01 |
ENST00000524506.5
ENST00000309657.8 |
RCE1
|
Ras converting CAAX endopeptidase 1 |
| chr19_-_38229654 | 0.01 |
ENST00000412732.5
ENST00000456296.5 |
DPF1
|
double PHD fingers 1 |
| chr7_-_156893150 | 0.01 |
ENST00000353442.10
|
LMBR1
|
limb development membrane protein 1 |
| chr19_+_40466976 | 0.01 |
ENST00000598249.6
|
SPTBN4
|
spectrin beta, non-erythrocytic 4 |
| chr19_+_40467145 | 0.01 |
ENST00000338932.7
|
SPTBN4
|
spectrin beta, non-erythrocytic 4 |
| chr3_-_52533776 | 0.01 |
ENST00000459839.5
|
NT5DC2
|
5'-nucleotidase domain containing 2 |
| chr9_-_132944600 | 0.01 |
ENST00000490179.3
ENST00000643583.1 ENST00000298552.9 ENST00000643072.1 ENST00000642745.1 ENST00000647462.1 ENST00000643875.1 ENST00000642627.1 ENST00000475903.6 ENST00000642617.1 ENST00000642646.1 ENST00000646625.1 ENST00000645150.1 ENST00000645129.1 ENST00000403810.6 ENST00000643691.1 ENST00000644097.1 |
TSC1
|
TSC complex subunit 1 |
| chr19_-_55117336 | 0.01 |
ENST00000592993.1
|
PPP1R12C
|
protein phosphatase 1 regulatory subunit 12C |
| chr14_-_54488940 | 0.01 |
ENST00000628554.2
ENST00000358056.8 |
GMFB
|
glia maturation factor beta |
| chr11_+_64286420 | 0.01 |
ENST00000313074.7
ENST00000542190.5 ENST00000541952.1 |
GPR137
|
G protein-coupled receptor 137 |
| chr6_-_36986122 | 0.01 |
ENST00000460219.2
ENST00000373627.10 ENST00000373616.9 |
MTCH1
|
mitochondrial carrier 1 |
| chr1_+_20290869 | 0.01 |
ENST00000289815.13
ENST00000375079.6 |
VWA5B1
|
von Willebrand factor A domain containing 5B1 |
| chr6_+_31762996 | 0.01 |
ENST00000415669.3
ENST00000425424.4 |
SAPCD1
|
suppressor APC domain containing 1 |
| chr10_-_70283998 | 0.01 |
ENST00000277942.7
|
NPFFR1
|
neuropeptide FF receptor 1 |
| chr2_-_109614143 | 0.01 |
ENST00000356688.8
|
SEPTIN10
|
septin 10 |
| chr19_-_49878350 | 0.01 |
ENST00000391834.6
ENST00000391830.1 |
AKT1S1
|
AKT1 substrate 1 |
| chr3_-_52533799 | 0.01 |
ENST00000422318.7
|
NT5DC2
|
5'-nucleotidase domain containing 2 |
| chr3_-_51967410 | 0.01 |
ENST00000461554.6
ENST00000483411.5 ENST00000461544.2 ENST00000355852.6 |
PCBP4
|
poly(rC) binding protein 4 |
| chr16_+_56451513 | 0.01 |
ENST00000562150.5
ENST00000561646.5 ENST00000566157.6 ENST00000568397.1 |
OGFOD1
|
2-oxoglutarate and iron dependent oxygenase domain containing 1 |
| chrX_+_119236245 | 0.01 |
ENST00000535419.2
|
PGRMC1
|
progesterone receptor membrane component 1 |
| chr1_-_151327650 | 0.01 |
ENST00000368875.6
ENST00000529142.5 |
PI4KB
|
phosphatidylinositol 4-kinase beta |
| chr1_-_92483947 | 0.01 |
ENST00000370332.5
|
GFI1
|
growth factor independent 1 transcriptional repressor |
| chrX_-_47145035 | 0.01 |
ENST00000276062.8
|
NDUFB11
|
NADH:ubiquinone oxidoreductase subunit B11 |
| chr2_-_55419565 | 0.01 |
ENST00000647341.1
ENST00000647401.1 ENST00000336838.10 ENST00000621814.4 ENST00000644033.1 ENST00000645477.1 ENST00000647517.1 |
CCDC88A
|
coiled-coil domain containing 88A |
| chr3_+_32685128 | 0.01 |
ENST00000331889.10
ENST00000328834.9 |
CNOT10
|
CCR4-NOT transcription complex subunit 10 |
| chr5_+_10353668 | 0.01 |
ENST00000274140.10
ENST00000449913.6 ENST00000503788.5 |
MARCHF6
|
membrane associated ring-CH-type finger 6 |
| chr19_-_43754901 | 0.01 |
ENST00000270066.11
ENST00000601170.5 |
SMG9
|
SMG9 nonsense mediated mRNA decay factor |
| chr21_-_14383125 | 0.01 |
ENST00000285667.4
|
HSPA13
|
heat shock protein family A (Hsp70) member 13 |
| chr1_-_151327684 | 0.01 |
ENST00000368874.8
|
PI4KB
|
phosphatidylinositol 4-kinase beta |
| chr17_+_50274447 | 0.01 |
ENST00000507382.2
|
TMEM92
|
transmembrane protein 92 |
| chr3_+_184135343 | 0.01 |
ENST00000648915.2
ENST00000432569.2 ENST00000647909.1 |
EIF2B5
|
eukaryotic translation initiation factor 2B subunit epsilon |
| chr10_-_73811583 | 0.01 |
ENST00000309979.11
|
NDST2
|
N-deacetylase and N-sulfotransferase 2 |
| chr14_+_79280056 | 0.01 |
ENST00000676811.1
|
NRXN3
|
neurexin 3 |
| chr6_+_29942523 | 0.01 |
ENST00000376809.10
ENST00000376802.2 ENST00000638375.1 |
HLA-A
|
major histocompatibility complex, class I, A |
| chr2_-_173964069 | 0.01 |
ENST00000652005.2
|
SP3
|
Sp3 transcription factor |
| chr16_+_32066065 | 0.01 |
ENST00000354689.6
|
IGHV3OR16-9
|
immunoglobulin heavy variable 3/OR16-9 (non-functional) |
| chr14_-_53153281 | 0.01 |
ENST00000357758.3
ENST00000673822.2 |
DDHD1
|
DDHD domain containing 1 |
| chr19_-_55117627 | 0.00 |
ENST00000263433.8
|
PPP1R12C
|
protein phosphatase 1 regulatory subunit 12C |
| chr2_-_173964180 | 0.00 |
ENST00000418194.7
|
SP3
|
Sp3 transcription factor |
Network of associatons between targets according to the STRING database.
First level regulatory network of PLAG1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.1 | 0.2 | GO:0072275 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.0 | 0.3 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 0.1 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.0 | 0.1 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.1 | GO:0061011 | hepatic duct development(GO:0061011) |
| 0.0 | 0.1 | GO:0072086 | specification of loop of Henle identity(GO:0072086) |
| 0.0 | 0.1 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.1 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) |
| 0.0 | 0.1 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.0 | 0.0 | GO:0044335 | canonical Wnt signaling pathway involved in neural crest cell differentiation(GO:0044335) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.2 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.0 | GO:0060559 | positive regulation of chronic inflammatory response to antigenic stimulus(GO:0002876) regulation of translational initiation by iron(GO:0006447) positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.0 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.0 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.0 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.2 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.2 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.0 | GO:1904928 | coreceptor activity involved in canonical Wnt signaling pathway(GO:1904928) |
| 0.0 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |