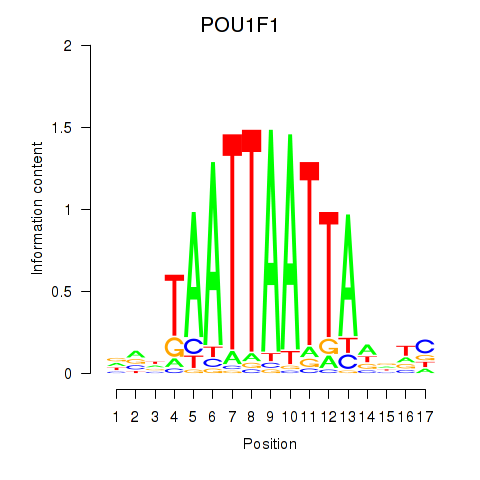
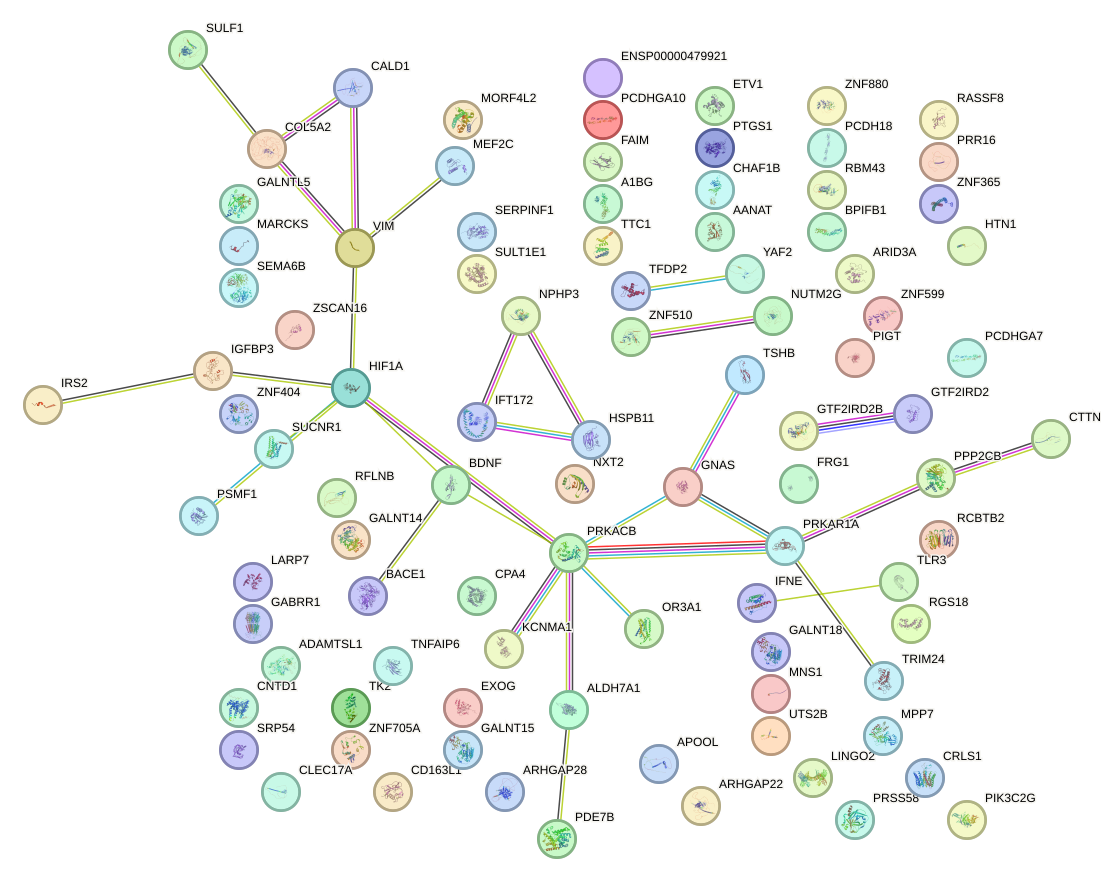
|
chr9_+_96928310
Show fit
|
0.45 |
ENST00000354649.7
|
NUTM2G
|
NUT family member 2G
|
|
chr5_+_120531464
Show fit
|
0.45 |
ENST00000505123.5
|
PRR16
|
proline rich 16
|
|
chr2_-_189179754
Show fit
|
0.37 |
ENST00000374866.9
ENST00000618828.1
|
COL5A2
|
collagen type V alpha 2 chain
|
|
chr4_+_70050431
Show fit
|
0.36 |
ENST00000511674.5
ENST00000246896.8
|
HTN1
|
histatin 1
|
|
chr19_+_49513353
Show fit
|
0.36 |
ENST00000596975.5
|
FCGRT
|
Fc fragment of IgG receptor and transporter
|
|
chr11_-_63608542
Show fit
|
0.32 |
ENST00000540943.1
|
PLAAT3
|
phospholipase A and acyltransferase 3
|
|
chr2_+_151357583
Show fit
|
0.30 |
ENST00000243347.5
|
TNFAIP6
|
TNF alpha induced protein 6
|
|
chr7_-_45921264
Show fit
|
0.30 |
ENST00000613132.5
ENST00000381083.9
ENST00000381086.9
|
IGFBP3
|
insulin like growth factor binding protein 3
|
|
chr8_+_69492793
Show fit
|
0.28 |
ENST00000616868.1
ENST00000419716.7
ENST00000402687.9
|
SULF1
|
sulfatase 1
|
|
chr5_-_88823763
Show fit
|
0.26 |
ENST00000635898.1
ENST00000626391.2
ENST00000628656.2
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr19_+_49513154
Show fit
|
0.26 |
ENST00000426395.7
ENST00000600273.5
ENST00000599988.5
|
FCGRT
|
Fc fragment of IgG receptor and transporter
|
|
chr5_+_141382702
Show fit
|
0.25 |
ENST00000617050.1
ENST00000518325.2
|
PCDHGA7
|
protocadherin gamma subfamily A, 7
|
|
chr19_-_58353482
Show fit
|
0.25 |
ENST00000263100.8
|
A1BG
|
alpha-1-B glycoprotein
|
|
chr7_+_130293134
Show fit
|
0.23 |
ENST00000445470.6
ENST00000492072.5
ENST00000222482.10
ENST00000473956.5
ENST00000493259.5
ENST00000486598.1
|
CPA4
|
carboxypeptidase A4
|
|
chr3_-_191282383
Show fit
|
0.21 |
ENST00000427544.6
|
UTS2B
|
urotensin 2B
|
|
chr12_+_18242955
Show fit
|
0.21 |
ENST00000676171.1
|
PIK3C2G
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma
|
|
chr17_+_1771688
Show fit
|
0.19 |
ENST00000572048.1
ENST00000573763.1
|
SERPINF1
|
serpin family F member 1
|
|
chr10_+_62374361
Show fit
|
0.17 |
ENST00000395254.8
|
ZNF365
|
zinc finger protein 365
|
|
chr10_-_48652493
Show fit
|
0.17 |
ENST00000435790.6
|
ARHGAP22
|
Rho GTPase activating protein 22
|
|
chr7_-_13986439
Show fit
|
0.16 |
ENST00000443608.5
ENST00000438956.5
|
ETV1
|
ETS variant transcription factor 1
|
|
chr10_-_77637789
Show fit
|
0.16 |
ENST00000481070.1
ENST00000640969.1
ENST00000286628.14
ENST00000638991.1
ENST00000639913.1
ENST00000480683.2
|
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1
|
|
chr10_-_77637721
Show fit
|
0.16 |
ENST00000638848.1
ENST00000639406.1
ENST00000618048.2
ENST00000639120.1
ENST00000640834.1
ENST00000639601.1
ENST00000638514.1
ENST00000457953.6
ENST00000639090.1
ENST00000639489.1
ENST00000372440.6
ENST00000404771.8
ENST00000638203.1
ENST00000638306.1
ENST00000638351.1
ENST00000638606.1
ENST00000639591.1
ENST00000640182.1
ENST00000640605.1
ENST00000640141.1
|
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1
|
|
chr16_-_66549839
Show fit
|
0.16 |
ENST00000527800.6
ENST00000677555.1
ENST00000563369.6
|
TK2
|
thymidine kinase 2
|
|
chr9_+_18474206
Show fit
|
0.16 |
ENST00000276935.6
|
ADAMTSL1
|
ADAMTS like 1
|
|
chr10_-_77637902
Show fit
|
0.15 |
ENST00000286627.10
ENST00000639486.1
ENST00000640523.1
|
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1
|
|
chr6_+_113857333
Show fit
|
0.15 |
ENST00000612661.2
|
MARCKS
|
myristoylated alanine rich protein kinase C substrate
|
|
chr16_-_66550091
Show fit
|
0.14 |
ENST00000564917.5
ENST00000677420.1
|
TK2
|
thymidine kinase 2
|
|
chr11_-_27700447
Show fit
|
0.14 |
ENST00000356660.9
|
BDNF
|
brain derived neurotrophic factor
|
|
chr16_-_66550005
Show fit
|
0.14 |
ENST00000527284.6
|
TK2
|
thymidine kinase 2
|
|
chr15_-_19988117
Show fit
|
0.14 |
ENST00000558565.2
|
IGHV3OR15-7
|
immunoglobulin heavy variable 3/OR15-7 (pseudogene)
|
|
chr7_+_134843884
Show fit
|
0.12 |
ENST00000445569.6
|
CALD1
|
caldesmon 1
|
|
chr1_-_53945584
Show fit
|
0.12 |
ENST00000371377.3
|
HSPB11
|
heat shock protein family B (small) member 11
|
|
chr1_-_53945567
Show fit
|
0.12 |
ENST00000371378.6
|
HSPB11
|
heat shock protein family B (small) member 11
|
|
chr10_+_87357720
Show fit
|
0.12 |
ENST00000412718.3
ENST00000381697.7
|
NUTM2D
|
NUT family member 2D
|
|
chr19_-_43883964
Show fit
|
0.11 |
ENST00000587539.2
|
ZNF404
|
zinc finger protein 404
|
|
chr10_-_27240505
Show fit
|
0.11 |
ENST00000375888.5
ENST00000676732.1
|
ACBD5
|
acyl-CoA binding domain containing 5
|
|
chr19_+_52369911
Show fit
|
0.11 |
ENST00000424032.6
ENST00000422689.3
ENST00000600321.5
ENST00000344085.9
ENST00000597976.5
|
ZNF880
|
zinc finger protein 880
|
|
chr9_-_96778053
Show fit
|
0.11 |
ENST00000375231.5
ENST00000223428.9
|
ZNF510
|
zinc finger protein 510
|
|
chr15_-_19965101
Show fit
|
0.11 |
ENST00000338912.5
|
IGHV1OR15-9
|
immunoglobulin heavy variable 1/OR15-9 (non-functional)
|
|
chr5_-_126595237
Show fit
|
0.11 |
ENST00000637206.1
ENST00000553117.5
|
ALDH7A1
|
aldehyde dehydrogenase 7 family member A1
|
|
chr1_+_84181630
Show fit
|
0.11 |
ENST00000610457.1
|
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta
|
|
chr5_+_141412979
Show fit
|
0.10 |
ENST00000612503.1
ENST00000398610.3
|
PCDHGA10
|
protocadherin gamma subfamily A, 10
|
|
chr1_+_115029823
Show fit
|
0.10 |
ENST00000256592.3
|
TSHB
|
thyroid stimulating hormone subunit beta
|
|
chr17_-_445939
Show fit
|
0.10 |
ENST00000329099.4
|
RFLNB
|
refilin B
|
|
chr10_+_17228215
Show fit
|
0.10 |
ENST00000544301.7
|
VIM
|
vimentin
|
|
chr18_+_6834473
Show fit
|
0.10 |
ENST00000581099.5
ENST00000419673.6
ENST00000531294.5
|
ARHGAP28
|
Rho GTPase activating protein 28
|
|
chr13_-_48533165
Show fit
|
0.09 |
ENST00000430805.6
ENST00000544492.5
ENST00000544904.3
|
RCBTB2
|
RCC1 and BTB domain containing protein 2
|
|
chr4_-_69860138
Show fit
|
0.09 |
ENST00000226444.4
|
SULT1E1
|
sulfotransferase family 1E member 1
|
|
chr3_+_138621225
Show fit
|
0.09 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule
|
|
chr1_-_53945661
Show fit
|
0.09 |
ENST00000194214.10
|
HSPB11
|
heat shock protein family B (small) member 11
|
|
chr2_+_161136901
Show fit
|
0.09 |
ENST00000259075.6
ENST00000432002.5
|
TANK
|
TRAF family member associated NFKB activator
|
|
chr16_-_66550142
Show fit
|
0.09 |
ENST00000417693.8
ENST00000299697.12
ENST00000451102.7
|
TK2
|
thymidine kinase 2
|
|
chr19_+_926001
Show fit
|
0.09 |
ENST00000263620.8
|
ARID3A
|
AT-rich interaction domain 3A
|
|
chr12_-_7444139
Show fit
|
0.09 |
ENST00000416109.2
ENST00000313599.8
|
CD163L1
|
CD163 molecule like 1
|
|
chr20_+_58907981
Show fit
|
0.09 |
ENST00000656419.1
|
GNAS
|
GNAS complex locus
|
|
chr14_+_34993240
Show fit
|
0.09 |
ENST00000677647.1
|
SRP54
|
signal recognition particle 54
|
|
chr20_+_1118590
Show fit
|
0.09 |
ENST00000246015.8
ENST00000335877.11
|
PSMF1
|
proteasome inhibitor subunit 1
|
|
chr20_+_6006039
Show fit
|
0.09 |
ENST00000452938.5
ENST00000378863.9
|
CRLS1
|
cardiolipin synthase 1
|
|
chr16_-_66550112
Show fit
|
0.09 |
ENST00000544898.6
ENST00000620035.5
ENST00000545043.6
|
TK2
|
thymidine kinase 2
|
|
chr12_+_8157034
Show fit
|
0.08 |
ENST00000396570.7
|
ZNF705A
|
zinc finger protein 705A
|
|
chr10_-_28282086
Show fit
|
0.08 |
ENST00000375719.7
ENST00000375732.5
|
MPP7
|
membrane palmitoylated protein 7
|
|
chr15_-_56465130
Show fit
|
0.08 |
ENST00000260453.4
|
MNS1
|
meiosis specific nuclear structural 1
|
|
chr6_+_28124596
Show fit
|
0.08 |
ENST00000340487.5
|
ZSCAN16
|
zinc finger and SCAN domain containing 16
|
|
chr5_+_160009113
Show fit
|
0.08 |
ENST00000522793.5
ENST00000682719.1
ENST00000684137.1
ENST00000683219.1
ENST00000684018.1
ENST00000231238.10
ENST00000682131.1
|
TTC1
|
tetratricopeptide repeat domain 1
|
|
chr4_+_186069144
Show fit
|
0.08 |
ENST00000513189.1
ENST00000296795.8
|
TLR3
|
toll like receptor 3
|
|
chr16_+_16340328
Show fit
|
0.07 |
ENST00000524823.6
|
ENSG00000183889.12
|
novel member of the nuclear pore complex interacting protein NPIP gene family
|
|
chr17_-_3292600
Show fit
|
0.07 |
ENST00000615105.1
|
OR3A1
|
olfactory receptor family 3 subfamily A member 1
|
|
chr19_-_34773184
Show fit
|
0.07 |
ENST00000588760.1
ENST00000329285.13
ENST00000587354.6
|
ZNF599
|
zinc finger protein 599
|
|
chrX_+_85003863
Show fit
|
0.07 |
ENST00000373173.7
|
APOOL
|
apolipoprotein O like
|
|
chr6_-_26234978
Show fit
|
0.07 |
ENST00000244534.7
|
H1-3
|
H1.3 linker histone, cluster member
|
|
chr12_+_26011713
Show fit
|
0.07 |
ENST00000542004.5
|
RASSF8
|
Ras association domain family member 8
|
|
chrX_-_103688033
Show fit
|
0.07 |
ENST00000434230.5
ENST00000418819.5
ENST00000360458.5
|
MORF4L2
|
mortality factor 4 like 2
|
|
chr7_-_142258027
Show fit
|
0.07 |
ENST00000552471.1
ENST00000547058.6
|
PRSS58
|
serine protease 58
|
|
chr17_+_68515399
Show fit
|
0.07 |
ENST00000588188.6
|
PRKAR1A
|
protein kinase cAMP-dependent type I regulatory subunit alpha
|
|
chr14_+_61697622
Show fit
|
0.07 |
ENST00000539097.2
|
HIF1A
|
hypoxia inducible factor 1 subunit alpha
|
|
chrX_+_109535775
Show fit
|
0.07 |
ENST00000218004.5
|
NXT2
|
nuclear transport factor 2 like export factor 2
|
|
chr9_+_122371014
Show fit
|
0.07 |
ENST00000362012.7
|
PTGS1
|
prostaglandin-endoperoxide synthase 1
|
|
chrX_-_103688090
Show fit
|
0.07 |
ENST00000433176.6
|
MORF4L2
|
mortality factor 4 like 2
|
|
chr14_-_106470788
Show fit
|
0.07 |
ENST00000434710.1
|
IGHV3-43
|
immunoglobulin heavy variable 3-43
|
|
chr2_-_151261839
Show fit
|
0.06 |
ENST00000331426.6
|
RBM43
|
RNA binding motif protein 43
|
|
chr7_+_75092573
Show fit
|
0.06 |
ENST00000651028.1
ENST00000472837.7
ENST00000629105.2
ENST00000619142.4
ENST00000614064.4
|
GTF2IRD2B
|
GTF2I repeat domain containing 2B
|
|
chr3_+_151873634
Show fit
|
0.06 |
ENST00000362032.6
|
SUCNR1
|
succinate receptor 1
|
|
chr9_-_21482313
Show fit
|
0.06 |
ENST00000448696.4
|
IFNE
|
interferon epsilon
|
|
chr7_-_13986498
Show fit
|
0.06 |
ENST00000420159.6
ENST00000399357.7
ENST00000403527.5
|
ETV1
|
ETS variant transcription factor 1
|
|
chr11_-_117316230
Show fit
|
0.06 |
ENST00000313005.11
ENST00000528053.5
|
BACE1
|
beta-secretase 1
|
|
chr19_+_14583076
Show fit
|
0.06 |
ENST00000547437.5
ENST00000417570.6
|
CLEC17A
|
C-type lectin domain containing 17A
|
|
chr4_-_137532452
Show fit
|
0.06 |
ENST00000412923.6
ENST00000511115.5
ENST00000344876.9
ENST00000507846.5
ENST00000510305.5
ENST00000611581.1
|
PCDH18
|
protocadherin 18
|
|
chr7_+_138460238
Show fit
|
0.06 |
ENST00000343526.9
|
TRIM24
|
tripartite motif containing 24
|
|
chr3_-_142000353
Show fit
|
0.06 |
ENST00000499676.5
|
TFDP2
|
transcription factor Dp-2
|
|
chr3_+_38496467
Show fit
|
0.06 |
ENST00000453767.1
|
EXOG
|
exo/endonuclease G
|
|
chr4_+_112647059
Show fit
|
0.06 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein 7, transcriptional regulator
|
|
chr7_-_74851518
Show fit
|
0.06 |
ENST00000651129.1
ENST00000614386.1
ENST00000625377.2
ENST00000451013.7
|
GTF2IRD2
|
GTF2I repeat domain containing 2
|
|
chr19_-_4558417
Show fit
|
0.06 |
ENST00000586965.1
|
SEMA6B
|
semaphorin 6B
|
|
chr3_-_132684685
Show fit
|
0.06 |
ENST00000512094.5
ENST00000632629.1
|
NPHP3
NPHP3-ACAD11
|
nephrocystin 3
NPHP3-ACAD11 readthrough (NMD candidate)
|
|
chr1_+_112396200
Show fit
|
0.06 |
ENST00000271277.11
|
CTTNBP2NL
|
CTTNBP2 N-terminal like
|
|
chr20_+_33283205
Show fit
|
0.06 |
ENST00000253354.2
|
BPIFB1
|
BPI fold containing family B member 1
|
|
chr14_-_106658251
Show fit
|
0.06 |
ENST00000454421.2
|
IGHV3-64
|
immunoglobulin heavy variable 3-64
|
|
chr12_-_42237727
Show fit
|
0.06 |
ENST00000548917.1
|
YAF2
|
YY1 associated factor 2
|
|
chr3_+_16174628
Show fit
|
0.06 |
ENST00000339732.10
|
GALNT15
|
polypeptide N-acetylgalactosaminyltransferase 15
|
|
chr1_+_8318088
Show fit
|
0.06 |
ENST00000471889.7
|
SLC45A1
|
solute carrier family 45 member 1
|
|
chr3_+_138621207
Show fit
|
0.05 |
ENST00000464668.5
|
FAIM
|
Fas apoptotic inhibitory molecule
|
|
chr17_+_76467597
Show fit
|
0.05 |
ENST00000392492.8
|
AANAT
|
aralkylamine N-acetyltransferase
|
|
chr2_-_27489716
Show fit
|
0.05 |
ENST00000260570.8
ENST00000675690.1
|
IFT172
|
intraflagellar transport 172
|
|
chr14_-_80231052
Show fit
|
0.05 |
ENST00000557010.5
|
DIO2
|
iodothyronine deiodinase 2
|
|
chr8_-_30812867
Show fit
|
0.05 |
ENST00000518243.5
|
PPP2CB
|
protein phosphatase 2 catalytic subunit beta
|
|
chr9_-_28670285
Show fit
|
0.05 |
ENST00000379992.6
ENST00000308675.5
ENST00000613945.3
|
LINGO2
|
leucine rich repeat and Ig domain containing 2
|
|
chr4_+_189940838
Show fit
|
0.05 |
ENST00000524583.5
ENST00000226798.9
ENST00000531991.6
|
FRG1
|
FSHD region gene 1
|
|
chr13_-_109786567
Show fit
|
0.05 |
ENST00000375856.5
|
IRS2
|
insulin receptor substrate 2
|
|
chr6_+_26103922
Show fit
|
0.05 |
ENST00000377803.4
|
H4C3
|
H4 clustered histone 3
|
|
chr6_+_135851681
Show fit
|
0.05 |
ENST00000308191.11
|
PDE7B
|
phosphodiesterase 7B
|
|
chr17_+_42798779
Show fit
|
0.05 |
ENST00000585355.5
|
CNTD1
|
cyclin N-terminal domain containing 1
|
|
chr1_+_192158448
Show fit
|
0.05 |
ENST00000367460.4
|
RGS18
|
regulator of G protein signaling 18
|
|
chr6_-_89217339
Show fit
|
0.05 |
ENST00000454853.7
|
GABRR1
|
gamma-aminobutyric acid type A receptor subunit rho1
|
|
chr20_+_45416551
Show fit
|
0.05 |
ENST00000639292.1
|
PIGT
|
phosphatidylinositol glycan anchor biosynthesis class T
|
|
chr20_+_6007245
Show fit
|
0.05 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1
|
|
chr7_-_122098831
Show fit
|
0.05 |
ENST00000681213.1
ENST00000679419.1
|
AASS
|
aminoadipate-semialdehyde synthase
|
|
chr7_-_22822829
Show fit
|
0.05 |
ENST00000358435.9
ENST00000621567.1
|
TOMM7
|
translocase of outer mitochondrial membrane 7
|
|
chr9_+_72577369
Show fit
|
0.05 |
ENST00000651183.1
|
TMC1
|
transmembrane channel like 1
|
|
chr6_+_158312459
Show fit
|
0.05 |
ENST00000367097.8
|
TULP4
|
TUB like protein 4
|
|
chr19_+_15082211
Show fit
|
0.05 |
ENST00000641398.1
|
OR1I1
|
olfactory receptor family 1 subfamily I member 1
|
|
chr19_-_14835252
Show fit
|
0.05 |
ENST00000641666.1
ENST00000642030.1
ENST00000642000.1
|
OR7C1
|
olfactory receptor family 7 subfamily C member 1
|
|
chr19_-_51417791
Show fit
|
0.05 |
ENST00000353836.9
|
SIGLEC10
|
sialic acid binding Ig like lectin 10
|
|
chr17_+_7407838
Show fit
|
0.05 |
ENST00000302926.7
|
NLGN2
|
neuroligin 2
|
|
chr5_+_154941063
Show fit
|
0.05 |
ENST00000523037.6
ENST00000265229.12
ENST00000439747.7
ENST00000522038.5
|
MRPL22
|
mitochondrial ribosomal protein L22
|
|
chr11_-_26572130
Show fit
|
0.05 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated
|
|
chr15_-_55917080
Show fit
|
0.04 |
ENST00000506154.1
|
NEDD4
|
NEDD4 E3 ubiquitin protein ligase
|
|
chr1_+_212791828
Show fit
|
0.04 |
ENST00000532324.5
ENST00000530441.5
ENST00000526641.5
ENST00000531963.5
ENST00000366973.8
ENST00000366974.9
ENST00000526997.5
ENST00000488246.6
|
TATDN3
|
TatD DNase domain containing 3
|
|
chr1_-_211134135
Show fit
|
0.04 |
ENST00000638983.1
ENST00000271751.10
ENST00000639952.1
|
ENSG00000284299.1
KCNH1
|
novel protein
potassium voltage-gated channel subfamily H member 1
|
|
chr10_-_6062349
Show fit
|
0.04 |
ENST00000379959.8
|
IL2RA
|
interleukin 2 receptor subunit alpha
|
|
chr10_+_70404129
Show fit
|
0.04 |
ENST00000373218.5
|
EIF4EBP2
|
eukaryotic translation initiation factor 4E binding protein 2
|
|
chr7_-_22822779
Show fit
|
0.04 |
ENST00000372879.8
|
TOMM7
|
translocase of outer mitochondrial membrane 7
|
|
chr19_+_4198075
Show fit
|
0.04 |
ENST00000262970.9
|
ANKRD24
|
ankyrin repeat domain 24
|
|
chr2_+_169584332
Show fit
|
0.04 |
ENST00000414307.6
ENST00000433207.6
ENST00000678088.1
ENST00000676508.1
ENST00000260970.8
ENST00000409714.7
|
PPIG
|
peptidylprolyl isomerase G
|
|
chr15_-_55917129
Show fit
|
0.04 |
ENST00000338963.6
ENST00000508342.5
|
NEDD4
|
NEDD4 E3 ubiquitin protein ligase
|
|
chr15_-_60397964
Show fit
|
0.04 |
ENST00000558998.5
ENST00000560165.5
ENST00000557986.5
ENST00000559467.5
ENST00000677968.1
ENST00000678450.1
ENST00000332680.8
ENST00000396024.7
ENST00000557906.6
ENST00000558558.6
ENST00000559113.6
ENST00000559780.6
ENST00000559956.6
ENST00000560468.6
ENST00000678870.1
ENST00000678061.1
ENST00000451270.7
ENST00000421017.6
ENST00000560466.5
ENST00000558132.5
ENST00000559370.5
ENST00000559725.5
ENST00000558985.6
ENST00000679109.1
|
ANXA2
|
annexin A2
|
|
chr11_+_110429898
Show fit
|
0.04 |
ENST00000260270.3
|
FDX1
|
ferredoxin 1
|
|
chr19_-_9107475
Show fit
|
0.04 |
ENST00000641081.1
|
OR7G2
|
olfactory receptor family 7 subfamily G member 2
|
|
chr20_-_31390580
Show fit
|
0.04 |
ENST00000339144.3
ENST00000376321.4
|
DEFB119
|
defensin beta 119
|
|
chr3_+_141738263
Show fit
|
0.04 |
ENST00000480908.1
ENST00000393000.3
ENST00000273480.4
|
RNF7
|
ring finger protein 7
|
|
chr9_+_72577788
Show fit
|
0.04 |
ENST00000645208.2
|
TMC1
|
transmembrane channel like 1
|
|
chr15_+_41286011
Show fit
|
0.04 |
ENST00000661438.1
|
ENSG00000285920.2
|
novel protein
|
|
chrX_+_56563569
Show fit
|
0.04 |
ENST00000338222.7
|
UBQLN2
|
ubiquilin 2
|
|
chr1_-_151831768
Show fit
|
0.04 |
ENST00000318247.7
|
RORC
|
RAR related orphan receptor C
|
|
chr6_+_47781982
Show fit
|
0.03 |
ENST00000489301.6
ENST00000638973.1
ENST00000371211.6
ENST00000393699.2
|
OPN5
|
opsin 5
|
|
chr12_+_130953898
Show fit
|
0.03 |
ENST00000261654.10
|
ADGRD1
|
adhesion G protein-coupled receptor D1
|
|
chr1_-_219613069
Show fit
|
0.03 |
ENST00000367211.6
ENST00000651890.1
|
ZC3H11B
|
zinc finger CCCH-type containing 11B
|
|
chr17_-_42798680
Show fit
|
0.03 |
ENST00000328434.8
|
COA3
|
cytochrome c oxidase assembly factor 3
|
|
chr11_-_63144221
Show fit
|
0.03 |
ENST00000417740.5
ENST00000612278.4
ENST00000326192.5
|
SLC22A24
|
solute carrier family 22 member 24
|
|
chr14_+_55027200
Show fit
|
0.03 |
ENST00000395472.2
ENST00000555846.2
|
SOCS4
|
suppressor of cytokine signaling 4
|
|
chr17_-_44066595
Show fit
|
0.03 |
ENST00000585388.2
ENST00000293406.8
|
LSM12
|
LSM12 homolog
|
|
chr9_+_108862255
Show fit
|
0.03 |
ENST00000333999.5
|
ACTL7A
|
actin like 7A
|
|
chr6_+_154039333
Show fit
|
0.03 |
ENST00000428397.6
|
OPRM1
|
opioid receptor mu 1
|
|
chr16_-_67183948
Show fit
|
0.03 |
ENST00000561621.5
ENST00000563902.2
ENST00000290881.11
|
KIAA0895L
|
KIAA0895 like
|
|
chr9_-_111330224
Show fit
|
0.03 |
ENST00000302681.3
|
OR2K2
|
olfactory receptor family 2 subfamily K member 2
|
|
chr19_-_44401575
Show fit
|
0.03 |
ENST00000591679.5
ENST00000544719.6
ENST00000614994.5
ENST00000585868.1
ENST00000588212.1
|
ZNF285
ENSG00000267173.1
|
zinc finger protein 285
novel protein
|
|
chr2_+_67397297
Show fit
|
0.03 |
ENST00000644028.1
ENST00000272342.6
|
ETAA1
|
ETAA1 activator of ATR kinase
|
|
chr6_+_29461440
Show fit
|
0.03 |
ENST00000396792.2
|
OR2H1
|
olfactory receptor family 2 subfamily H member 1
|
|
chr6_-_25874212
Show fit
|
0.03 |
ENST00000361703.10
ENST00000397060.8
|
SLC17A3
|
solute carrier family 17 member 3
|
|
chr13_+_30422487
Show fit
|
0.03 |
ENST00000638137.1
ENST00000635918.2
|
UBE2L5
|
ubiquitin conjugating enzyme E2 L5
|
|
chr1_-_53142617
Show fit
|
0.03 |
ENST00000371491.4
ENST00000371494.9
|
SLC1A7
|
solute carrier family 1 member 7
|
|
chr8_-_100105619
Show fit
|
0.03 |
ENST00000523287.5
ENST00000519092.5
|
RGS22
|
regulator of G protein signaling 22
|
|
chr1_-_212791762
Show fit
|
0.03 |
ENST00000626725.1
ENST00000366977.8
ENST00000366976.3
|
NSL1
|
NSL1 component of MIS12 kinetochore complex
|
|
chr14_+_21918161
Show fit
|
0.03 |
ENST00000390439.2
|
TRAV13-2
|
T cell receptor alpha variable 13-2
|
|
chr14_-_89954518
Show fit
|
0.03 |
ENST00000556005.1
ENST00000555872.5
|
EFCAB11
|
EF-hand calcium binding domain 11
|
|
chrX_-_13817027
Show fit
|
0.03 |
ENST00000493677.5
ENST00000355135.6
ENST00000316715.9
|
GPM6B
|
glycoprotein M6B
|
|
chr5_+_151020438
Show fit
|
0.03 |
ENST00000622181.4
ENST00000614343.4
ENST00000388825.9
ENST00000521650.5
ENST00000517973.1
|
GPX3
|
glutathione peroxidase 3
|
|
chr4_-_117085541
Show fit
|
0.03 |
ENST00000310754.5
|
TRAM1L1
|
translocation associated membrane protein 1 like 1
|
|
chr1_+_197268222
Show fit
|
0.03 |
ENST00000367400.8
ENST00000638467.1
ENST00000367399.6
|
CRB1
|
crumbs cell polarity complex component 1
|
|
chr14_+_55027576
Show fit
|
0.03 |
ENST00000339298.2
|
SOCS4
|
suppressor of cytokine signaling 4
|
|
chr15_+_64094060
Show fit
|
0.03 |
ENST00000560829.5
|
SNX1
|
sorting nexin 1
|
|
chr15_-_75455767
Show fit
|
0.03 |
ENST00000360439.8
|
SIN3A
|
SIN3 transcription regulator family member A
|
|
chr10_+_18260715
Show fit
|
0.03 |
ENST00000615785.4
ENST00000617363.4
ENST00000396576.6
|
CACNB2
|
calcium voltage-gated channel auxiliary subunit beta 2
|
|
chr1_-_53142577
Show fit
|
0.03 |
ENST00000620347.5
ENST00000611397.5
|
SLC1A7
|
solute carrier family 1 member 7
|
|
chr18_+_34976928
Show fit
|
0.03 |
ENST00000591734.5
ENST00000413393.5
ENST00000589180.5
ENST00000587359.5
|
MAPRE2
|
microtubule associated protein RP/EB family member 2
|
|
chr3_-_157503574
Show fit
|
0.03 |
ENST00000494677.5
ENST00000468233.5
|
VEPH1
|
ventricular zone expressed PH domain containing 1
|
|
chr1_-_205121964
Show fit
|
0.03 |
ENST00000264515.11
|
RBBP5
|
RB binding protein 5, histone lysine methyltransferase complex subunit
|
|
chr14_-_89954659
Show fit
|
0.03 |
ENST00000555070.1
ENST00000316738.12
ENST00000538485.6
ENST00000556609.5
|
ENSG00000259053.1
EFCAB11
|
novel transcript
EF-hand calcium binding domain 11
|
|
chr14_+_19743571
Show fit
|
0.03 |
ENST00000642117.2
|
OR4Q3
|
olfactory receptor family 4 subfamily Q member 3
|
|
chr7_-_87713287
Show fit
|
0.03 |
ENST00000416177.1
ENST00000265724.8
ENST00000543898.5
|
ABCB1
|
ATP binding cassette subfamily B member 1
|
|
chr6_+_29111560
Show fit
|
0.03 |
ENST00000377169.2
|
OR2J3
|
olfactory receptor family 2 subfamily J member 3
|
|
chr12_+_92702843
Show fit
|
0.03 |
ENST00000397833.3
|
PLEKHG7
|
pleckstrin homology and RhoGEF domain containing G7
|
|
chr9_-_113303271
Show fit
|
0.02 |
ENST00000297894.5
ENST00000489339.2
|
RNF183
|
ring finger protein 183
|
|
chr18_+_616672
Show fit
|
0.02 |
ENST00000338387.11
|
CLUL1
|
clusterin like 1
|
|
chr11_+_59511368
Show fit
|
0.02 |
ENST00000641278.1
|
OR4D9
|
olfactory receptor family 4 subfamily D member 9
|
|
chr15_-_60398733
Show fit
|
0.02 |
ENST00000559818.6
|
ANXA2
|
annexin A2
|
|
chr12_-_10130082
Show fit
|
0.02 |
ENST00000533022.5
|
CLEC7A
|
C-type lectin domain containing 7A
|
|
chr11_-_26572254
Show fit
|
0.02 |
ENST00000529533.6
|
MUC15
|
mucin 15, cell surface associated
|
|
chr19_-_22010930
Show fit
|
0.02 |
ENST00000601773.5
ENST00000397126.9
ENST00000609966.5
ENST00000601993.1
ENST00000599916.5
|
ZNF208
|
zinc finger protein 208
|
|
chr3_-_108757407
Show fit
|
0.02 |
ENST00000295755.7
|
RETNLB
|
resistin like beta
|
|
chr10_+_92593112
Show fit
|
0.02 |
ENST00000260731.5
|
KIF11
|
kinesin family member 11
|
|
chr8_+_36784324
Show fit
|
0.02 |
ENST00000523973.5
ENST00000399881.8
|
KCNU1
|
potassium calcium-activated channel subfamily U member 1
|
|
chr7_-_77199808
Show fit
|
0.02 |
ENST00000248598.6
|
FGL2
|
fibrinogen like 2
|
|
chr9_-_34729482
Show fit
|
0.02 |
ENST00000378788.4
|
FAM205A
|
family with sequence similarity 205 member A
|
|
chr4_-_121164314
Show fit
|
0.02 |
ENST00000057513.8
|
TNIP3
|
TNFAIP3 interacting protein 3
|
|
chrX_-_103686687
Show fit
|
0.02 |
ENST00000441076.7
ENST00000422355.5
ENST00000442614.5
ENST00000451301.5
|
MORF4L2
|
mortality factor 4 like 2
|
|
chr5_+_129748091
Show fit
|
0.02 |
ENST00000564719.2
|
MINAR2
|
membrane integral NOTCH2 associated receptor 2
|
|
chr5_-_135399863
Show fit
|
0.02 |
ENST00000510038.1
ENST00000304332.8
|
MACROH2A1
|
macroH2A.1 histone
|
|
chrX_+_71283577
Show fit
|
0.02 |
ENST00000420903.6
ENST00000373856.8
ENST00000678437.1
ENST00000678830.1
ENST00000677879.1
ENST00000373841.5
ENST00000276079.13
ENST00000676797.1
ENST00000678660.1
ENST00000678231.1
ENST00000677612.1
ENST00000413858.5
ENST00000450092.6
|
NONO
|
non-POU domain containing octamer binding
|
|
chr1_+_152970646
Show fit
|
0.02 |
ENST00000328051.3
|
SPRR4
|
small proline rich protein 4
|
|
chrX_+_136648138
Show fit
|
0.02 |
ENST00000370629.7
|
CD40LG
|
CD40 ligand
|