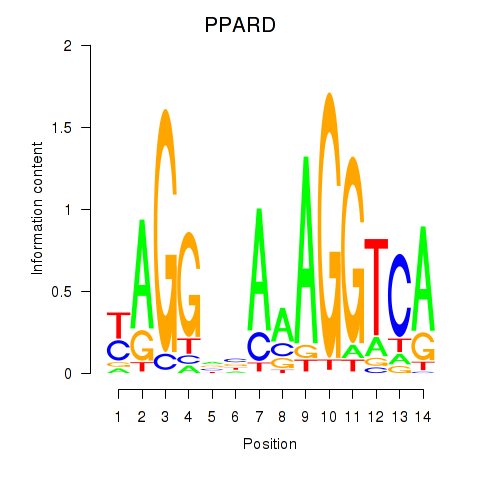
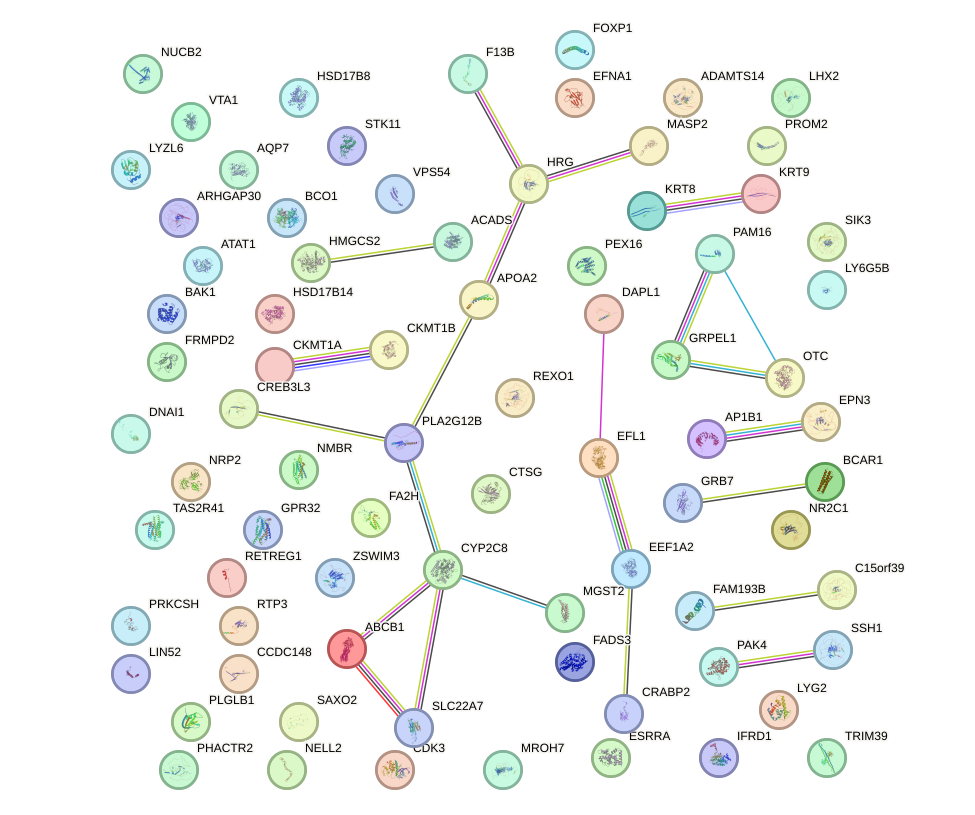
|
chr15_+_43593054
Show fit
|
0.52 |
ENST00000453782.5
ENST00000300283.10
ENST00000437924.5
|
CKMT1B
|
creatine kinase, mitochondrial 1B
|
|
chr15_+_43692886
Show fit
|
0.48 |
ENST00000434505.5
ENST00000411750.5
|
CKMT1A
|
creatine kinase, mitochondrial 1A
|
|
chr19_-_10577231
Show fit
|
0.30 |
ENST00000589348.1
ENST00000592285.1
ENST00000587069.5
|
AP1M2
|
adaptor related protein complex 1 subunit mu 2
|
|
chr17_+_50532713
Show fit
|
0.26 |
ENST00000503690.5
ENST00000514874.5
ENST00000268933.8
|
EPN3
|
epsin 3
|
|
chr2_+_205682491
Show fit
|
0.22 |
ENST00000360409.7
ENST00000450507.5
ENST00000357785.10
ENST00000417189.5
|
NRP2
|
neuropilin 2
|
|
chr5_-_16508858
Show fit
|
0.15 |
ENST00000684456.1
|
RETREG1
|
reticulophagy regulator 1
|
|
chr5_-_16508951
Show fit
|
0.15 |
ENST00000682628.1
|
RETREG1
|
reticulophagy regulator 1
|
|
chr12_-_52926459
Show fit
|
0.15 |
ENST00000552150.5
|
KRT8
|
keratin 8
|
|
chr5_-_16508812
Show fit
|
0.14 |
ENST00000683414.1
|
RETREG1
|
reticulophagy regulator 1
|
|
chr1_+_61952283
Show fit
|
0.14 |
ENST00000307297.8
|
PATJ
|
PATJ crumbs cell polarity complex component
|
|
chr1_+_61952036
Show fit
|
0.14 |
ENST00000646453.1
ENST00000635137.1
|
PATJ
|
PATJ crumbs cell polarity complex component
|
|
chr4_+_139665768
Show fit
|
0.13 |
ENST00000616265.4
ENST00000265498.6
ENST00000506797.5
|
MGST2
|
microsomal glutathione S-transferase 2
|
|
chr5_-_16508990
Show fit
|
0.13 |
ENST00000399793.6
|
RETREG1
|
reticulophagy regulator 1
|
|
chr5_-_16508788
Show fit
|
0.13 |
ENST00000682142.1
|
RETREG1
|
reticulophagy regulator 1
|
|
chr1_-_156705764
Show fit
|
0.13 |
ENST00000621784.4
ENST00000368220.1
|
CRABP2
|
cellular retinoic acid binding protein 2
|
|
chr2_+_95274439
Show fit
|
0.12 |
ENST00000317620.14
ENST00000403131.6
ENST00000317668.8
|
PROM2
|
prominin 2
|
|
chr3_+_50273119
Show fit
|
0.12 |
ENST00000456560.6
ENST00000418576.3
|
SEMA3B
|
semaphorin 3B
|
|
chr1_+_6451304
Show fit
|
0.11 |
ENST00000636644.1
|
ESPN
|
espin
|
|
chr17_-_41572052
Show fit
|
0.09 |
ENST00000588431.1
ENST00000246662.9
|
KRT9
|
keratin 9
|
|
chr12_-_95073490
Show fit
|
0.07 |
ENST00000330677.7
|
NR2C1
|
nuclear receptor subfamily 2 group C member 1
|
|
chr15_+_75201873
Show fit
|
0.07 |
ENST00000394987.5
|
C15orf39
|
chromosome 15 open reading frame 39
|
|
chr17_+_39737923
Show fit
|
0.07 |
ENST00000577695.5
ENST00000309156.9
|
GRB7
|
growth factor receptor bound protein 7
|
|
chr9_-_33402551
Show fit
|
0.06 |
ENST00000297988.6
ENST00000624075.3
ENST00000625032.1
ENST00000625109.3
|
AQP7
|
aquaporin 7
|
|
chr7_+_143477873
Show fit
|
0.06 |
ENST00000408916.1
|
TAS2R41
|
taste 2 receptor member 41
|
|
chr19_-_1822038
Show fit
|
0.06 |
ENST00000643515.1
|
REXO1
|
RNA exonuclease 1 homolog
|
|
chr1_+_202348727
Show fit
|
0.06 |
ENST00000356764.6
|
PPP1R12B
|
protein phosphatase 1 regulatory subunit 12B
|
|
chr6_+_33204645
Show fit
|
0.06 |
ENST00000374662.4
|
HSD17B8
|
hydroxysteroid 17-beta dehydrogenase 8
|
|
chr9_-_33402449
Show fit
|
0.05 |
ENST00000377425.8
|
AQP7
|
aquaporin 7
|
|
chr7_+_112450451
Show fit
|
0.05 |
ENST00000429071.5
ENST00000403825.8
ENST00000675268.1
|
IFRD1
ENSG00000288634.1
|
interferon related developmental regulator 1
novel protein
|
|
chr22_-_42130800
Show fit
|
0.05 |
ENST00000645361.2
ENST00000359033.4
|
CYP2D6
|
cytochrome P450 family 2 subfamily D member 6
|
|
chr11_-_61891381
Show fit
|
0.05 |
ENST00000525588.5
|
FADS3
|
fatty acid desaturase 3
|
|
chr1_-_119768892
Show fit
|
0.05 |
ENST00000369406.8
ENST00000544913.2
|
HMGCS2
|
3-hydroxy-3-methylglutaryl-CoA synthase 2
|
|
chr19_+_11435619
Show fit
|
0.05 |
ENST00000589126.5
ENST00000588269.1
ENST00000587509.5
ENST00000591462.6
ENST00000592741.5
ENST00000677123.1
ENST00000593101.5
ENST00000587327.5
|
PRKCSH
|
protein kinase C substrate 80K-H
|
|
chr2_-_99255107
Show fit
|
0.04 |
ENST00000333017.6
ENST00000626374.2
ENST00000409679.5
ENST00000423306.1
|
LYG2
|
lysozyme g2
|
|
chr11_-_117098415
Show fit
|
0.04 |
ENST00000445177.6
ENST00000375300.6
ENST00000446921.6
|
SIK3
|
SIK family kinase 3
|
|
chr1_-_161069857
Show fit
|
0.04 |
ENST00000368013.8
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr14_+_74084947
Show fit
|
0.04 |
ENST00000674221.1
ENST00000554938.2
|
LIN52
|
lin-52 DREAM MuvB core complex component
|
|
chr16_+_81238682
Show fit
|
0.04 |
ENST00000258168.7
ENST00000564552.1
|
BCO1
|
beta-carotene oxygenase 1
|
|
chr2_+_158795309
Show fit
|
0.04 |
ENST00000309950.8
ENST00000621326.4
ENST00000409042.5
|
DAPL1
|
death associated protein like 1
|
|
chr11_-_61891534
Show fit
|
0.04 |
ENST00000278829.7
|
FADS3
|
fatty acid desaturase 3
|
|
chr1_-_161069962
Show fit
|
0.04 |
ENST00000368015.1
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr2_-_64019367
Show fit
|
0.04 |
ENST00000272322.9
|
VPS54
|
VPS54 subunit of GARP complex
|
|
chr19_+_1205761
Show fit
|
0.04 |
ENST00000326873.12
ENST00000586243.5
|
STK11
|
serine/threonine kinase 11
|
|
chr16_-_4351283
Show fit
|
0.03 |
ENST00000318059.8
|
PAM16
|
presequence translocase associated motor 16
|
|
chr22_-_29388530
Show fit
|
0.03 |
ENST00000357586.7
ENST00000432560.6
ENST00000405198.6
ENST00000317368.11
|
AP1B1
|
adaptor related protein complex 1 subunit beta 1
|
|
chr2_+_218568809
Show fit
|
0.03 |
ENST00000273064.11
|
CNOT9
|
CCR4-NOT transcription complex subunit 9
|
|
chr2_+_218568865
Show fit
|
0.03 |
ENST00000295701.9
|
CNOT9
|
CCR4-NOT transcription complex subunit 9
|
|
chr6_+_43298326
Show fit
|
0.03 |
ENST00000372574.7
|
SLC22A7
|
solute carrier family 22 member 7
|
|
chr11_-_45917823
Show fit
|
0.03 |
ENST00000533151.5
ENST00000241041.7
|
PEX16
|
peroxisomal biogenesis factor 16
|
|
chr16_-_4351257
Show fit
|
0.03 |
ENST00000577031.5
|
PAM16
|
presequence translocase associated motor 16
|
|
chr10_-_48251757
Show fit
|
0.03 |
ENST00000305531.3
|
FRMPD2
|
FERM and PDZ domain containing 2
|
|
chr10_+_70672457
Show fit
|
0.03 |
ENST00000373207.2
|
ADAMTS14
|
ADAM metallopeptidase with thrombospondin type 1 motif 14
|
|
chr16_-_75251482
Show fit
|
0.03 |
ENST00000393420.10
ENST00000162330.10
|
BCAR1
|
BCAR1 scaffold protein, Cas family member
|
|
chr19_+_39125769
Show fit
|
0.03 |
ENST00000602004.1
ENST00000599470.5
ENST00000321944.8
ENST00000593480.5
ENST00000358301.7
ENST00000593690.5
ENST00000599386.5
|
PAK4
|
p21 (RAC1) activated kinase 4
|
|
chr4_-_7068033
Show fit
|
0.03 |
ENST00000264954.5
|
GRPEL1
|
GrpE like 1, mitochondrial
|
|
chr6_+_30327259
Show fit
|
0.03 |
ENST00000376659.9
ENST00000428555.1
|
TRIM39
|
tripartite motif containing 39
|
|
chr15_-_82262660
Show fit
|
0.03 |
ENST00000557844.1
ENST00000359445.7
ENST00000268206.12
|
EFL1
|
elongation factor like GTPase 1
|
|
chr14_-_24576240
Show fit
|
0.03 |
ENST00000216336.3
|
CTSG
|
cathepsin G
|
|
chr6_+_32038382
Show fit
|
0.02 |
ENST00000478281.5
ENST00000471671.4
ENST00000435122.3
ENST00000644719.2
|
CYP21A2
|
cytochrome P450 family 21 subfamily A member 2
|
|
chr1_-_11047225
Show fit
|
0.02 |
ENST00000400898.3
ENST00000400897.8
|
MASP2
|
mannan binding lectin serine peptidase 2
|
|
chr20_+_45857607
Show fit
|
0.02 |
ENST00000255152.3
|
ZSWIM3
|
zinc finger SWIM-type containing 3
|
|
chr6_-_142147122
Show fit
|
0.02 |
ENST00000258042.2
|
NMBR
|
neuromedin B receptor
|
|
chr2_-_158456702
Show fit
|
0.02 |
ENST00000409889.1
ENST00000283233.10
|
CCDC148
|
coiled-coil domain containing 148
|
|
chr17_-_35943707
Show fit
|
0.02 |
ENST00000615905.5
|
LYZL6
|
lysozyme like 6
|
|
chr19_+_11436044
Show fit
|
0.02 |
ENST00000589838.5
|
PRKCSH
|
protein kinase C substrate 80K-H
|
|
chr17_-_35943662
Show fit
|
0.02 |
ENST00000618542.4
|
LYZL6
|
lysozyme like 6
|
|
chr19_-_48835823
Show fit
|
0.02 |
ENST00000595764.1
|
HSD17B14
|
hydroxysteroid 17-beta dehydrogenase 14
|
|
chr12_-_108826161
Show fit
|
0.02 |
ENST00000546697.1
|
SSH1
|
slingshot protein phosphatase 1
|
|
chr1_+_54641806
Show fit
|
0.02 |
ENST00000409996.5
|
MROH7
|
maestro heat like repeat family member 7
|
|
chr15_+_82262781
Show fit
|
0.02 |
ENST00000566205.1
ENST00000339465.5
ENST00000569120.1
ENST00000682753.1
ENST00000566861.5
ENST00000565432.1
|
SAXO2
|
stabilizer of axonemal microtubules 2
|
|
chr1_-_197067234
Show fit
|
0.02 |
ENST00000367412.2
|
F13B
|
coagulation factor XIII B chain
|
|
chr22_+_22822658
Show fit
|
0.02 |
ENST00000620395.2
|
IGLV2-8
|
immunoglobulin lambda variable 2-8
|
|
chr19_+_4153616
Show fit
|
0.02 |
ENST00000078445.7
ENST00000595923.5
ENST00000602257.5
ENST00000602147.1
|
CREB3L3
|
cAMP responsive element binding protein 3 like 3
|
|
chr10_-_95069489
Show fit
|
0.02 |
ENST00000371270.6
ENST00000535898.5
ENST00000623108.3
|
CYP2C8
|
cytochrome P450 family 2 subfamily C member 8
|
|
chr6_+_30626842
Show fit
|
0.02 |
ENST00000318999.11
ENST00000376485.8
ENST00000330083.6
ENST00000319027.9
ENST00000376483.8
ENST00000329992.12
|
ATAT1
|
alpha tubulin acetyltransferase 1
|
|
chr1_-_161223559
Show fit
|
0.02 |
ENST00000469730.2
ENST00000463273.5
ENST00000464492.5
ENST00000367990.7
ENST00000470459.6
ENST00000463812.1
ENST00000468465.5
|
APOA2
|
apolipoprotein A2
|
|
chr6_+_143677935
Show fit
|
0.02 |
ENST00000440869.6
ENST00000367582.7
ENST00000451827.6
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr1_+_54641754
Show fit
|
0.01 |
ENST00000339553.9
ENST00000421030.7
|
MROH7
|
maestro heat like repeat family member 7
|
|
chr2_+_206159580
Show fit
|
0.01 |
ENST00000236957.9
ENST00000392221.5
ENST00000445505.5
|
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2
|
|
chr9_+_124011738
Show fit
|
0.01 |
ENST00000373615.9
|
LHX2
|
LIM homeobox 2
|
|
chr19_+_50770464
Show fit
|
0.01 |
ENST00000270590.4
|
GPR32
|
G protein-coupled receptor 32
|
|
chr11_-_45917867
Show fit
|
0.01 |
ENST00000378750.10
|
PEX16
|
peroxisomal biogenesis factor 16
|
|
chr20_-_63499074
Show fit
|
0.01 |
ENST00000217182.6
ENST00000642899.1
|
EEF1A2
|
eukaryotic translation elongation factor 1 alpha 2
|
|
chr1_-_161069666
Show fit
|
0.01 |
ENST00000368016.7
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr2_-_197499826
Show fit
|
0.01 |
ENST00000439605.2
ENST00000388968.8
ENST00000418022.2
|
HSPD1
|
heat shock protein family D (Hsp60) member 1
|
|
chr16_-_74774812
Show fit
|
0.01 |
ENST00000219368.8
|
FA2H
|
fatty acid 2-hydroxylase
|
|
chr1_-_145707387
Show fit
|
0.01 |
ENST00000451928.6
|
PDZK1
|
PDZ domain containing 1
|
|
chr12_+_130953898
Show fit
|
0.01 |
ENST00000261654.10
|
ADGRD1
|
adhesion G protein-coupled receptor D1
|
|
chr1_-_145707345
Show fit
|
0.01 |
ENST00000417171.6
|
PDZK1
|
PDZ domain containing 1
|
|
chr10_-_72954790
Show fit
|
0.01 |
ENST00000373032.4
|
PLA2G12B
|
phospholipase A2 group XIIB
|
|
chr2_-_197499857
Show fit
|
0.01 |
ENST00000428204.6
ENST00000678170.1
ENST00000676933.1
ENST00000678621.1
|
HSPD1
|
heat shock protein family D (Hsp60) member 1
|
|
chr11_+_64306227
Show fit
|
0.01 |
ENST00000405666.5
ENST00000468670.2
|
ESRRA
|
estrogen related receptor alpha
|
|
chr2_+_206159884
Show fit
|
0.01 |
ENST00000392222.7
|
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2
|
|
chr7_-_87713287
Show fit
|
0.01 |
ENST00000416177.1
ENST00000265724.8
ENST00000543898.5
|
ABCB1
|
ATP binding cassette subfamily B member 1
|
|
chr6_+_31670167
Show fit
|
0.01 |
ENST00000375864.4
|
LY6G5B
|
lymphocyte antigen 6 family member G5B
|
|
chr11_+_17260353
Show fit
|
0.01 |
ENST00000530527.5
|
NUCB2
|
nucleobindin 2
|
|
chr6_-_33580229
Show fit
|
0.01 |
ENST00000374467.4
ENST00000442998.6
ENST00000360661.9
|
BAK1
|
BCL2 antagonist/killer 1
|
|
chr2_+_218568558
Show fit
|
0.01 |
ENST00000627282.2
ENST00000542068.5
|
CNOT9
|
CCR4-NOT transcription complex subunit 9
|
|
chr1_+_6451578
Show fit
|
0.00 |
ENST00000434576.2
ENST00000477679.2
|
ESPN
|
espin
|
|
chr3_-_71132099
Show fit
|
0.00 |
ENST00000650188.1
ENST00000648121.1
ENST00000648794.1
ENST00000649592.1
|
FOXP1
|
forkhead box P1
|
|
chr17_+_76000906
Show fit
|
0.00 |
ENST00000448471.2
|
CDK3
|
cyclin dependent kinase 3
|
|
chr19_+_10086305
Show fit
|
0.00 |
ENST00000253110.16
ENST00000591813.5
|
SHFL
|
shiftless antiviral inhibitor of ribosomal frameshifting
|
|
chr3_+_46497970
Show fit
|
0.00 |
ENST00000296142.4
|
RTP3
|
receptor transporter protein 3
|
|
chr3_+_186666003
Show fit
|
0.00 |
ENST00000232003.5
|
HRG
|
histidine rich glycoprotein
|
|
chr20_-_775979
Show fit
|
0.00 |
ENST00000676154.1
ENST00000674666.1
ENST00000217254.11
|
SLC52A3
|
solute carrier family 52 member 3
|
|
chr12_+_120725796
Show fit
|
0.00 |
ENST00000242592.9
ENST00000411593.2
|
ACADS
|
acyl-CoA dehydrogenase short chain
|
|
chr6_+_142147162
Show fit
|
0.00 |
ENST00000452973.6
ENST00000620996.4
ENST00000367621.1
ENST00000367630.9
|
VTA1
|
vesicle trafficking 1
|
|
chr5_-_177554545
Show fit
|
0.00 |
ENST00000514747.6
|
FAM193B
|
family with sequence similarity 193 member B
|
|
chrX_+_38352573
Show fit
|
0.00 |
ENST00000039007.5
|
OTC
|
ornithine transcarbamylase
|
|
chr9_+_34458752
Show fit
|
0.00 |
ENST00000614641.4
ENST00000242317.9
ENST00000437363.5
|
DNAI1
|
dynein axonemal intermediate chain 1
|