Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
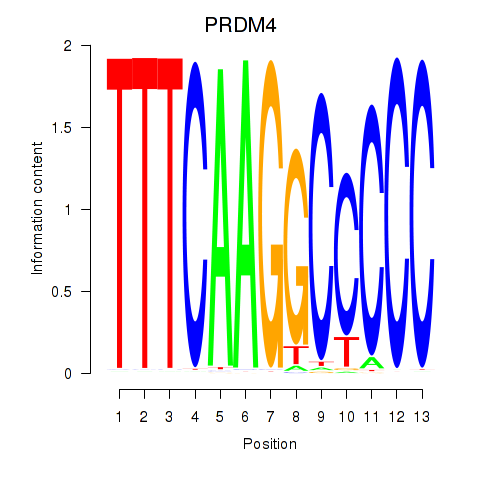
Results for PRDM4
Z-value: 0.14
Transcription factors associated with PRDM4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PRDM4
|
ENSG00000110851.12 | PR/SET domain 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PRDM4 | hg38_v1_chr12_-_107761113_107761176 | -0.76 | 2.9e-02 | Click! |
Activity profile of PRDM4 motif
Sorted Z-values of PRDM4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_35085295 | 0.12 |
ENST00000528386.4
|
NANOGP8
|
Nanog homeobox retrogene P8 |
| chr12_-_57846686 | 0.11 |
ENST00000548823.1
ENST00000398073.7 |
CTDSP2
|
CTD small phosphatase 2 |
| chr4_+_673897 | 0.09 |
ENST00000505477.5
|
MYL5
|
myosin light chain 5 |
| chr19_-_48868590 | 0.09 |
ENST00000263265.11
|
PLEKHA4
|
pleckstrin homology domain containing A4 |
| chr4_+_673518 | 0.09 |
ENST00000506838.5
|
MYL5
|
myosin light chain 5 |
| chr19_-_48868454 | 0.08 |
ENST00000355496.9
|
PLEKHA4
|
pleckstrin homology domain containing A4 |
| chr20_+_64066211 | 0.08 |
ENST00000458442.1
|
TCEA2
|
transcription elongation factor A2 |
| chr1_-_68050615 | 0.07 |
ENST00000646789.1
|
DIRAS3
|
DIRAS family GTPase 3 |
| chr1_+_42767241 | 0.07 |
ENST00000372525.7
|
C1orf50
|
chromosome 1 open reading frame 50 |
| chr12_-_10723307 | 0.06 |
ENST00000279550.11
ENST00000228251.9 |
YBX3
|
Y-box binding protein 3 |
| chr11_+_47257953 | 0.06 |
ENST00000437276.1
ENST00000436029.5 ENST00000467728.5 ENST00000441012.7 ENST00000405853.7 |
NR1H3
|
nuclear receptor subfamily 1 group H member 3 |
| chr10_-_103918128 | 0.06 |
ENST00000224950.8
|
STN1
|
STN1 subunit of CST complex |
| chr10_-_103452384 | 0.06 |
ENST00000369788.7
|
CALHM2
|
calcium homeostasis modulator family member 2 |
| chr20_+_62938111 | 0.05 |
ENST00000266069.5
|
GID8
|
GID complex subunit 8 homolog |
| chr12_+_15322529 | 0.05 |
ENST00000348962.7
|
PTPRO
|
protein tyrosine phosphatase receptor type O |
| chr16_-_67936808 | 0.05 |
ENST00000358514.9
|
PSMB10
|
proteasome 20S subunit beta 10 |
| chr20_+_59721210 | 0.05 |
ENST00000395636.6
ENST00000361300.4 |
PHACTR3
|
phosphatase and actin regulator 3 |
| chr10_+_86968432 | 0.05 |
ENST00000416348.1
ENST00000372013.8 |
ADIRF
|
adipogenesis regulatory factor |
| chr9_-_124500986 | 0.04 |
ENST00000373587.3
|
NR5A1
|
nuclear receptor subfamily 5 group A member 1 |
| chr19_-_11529094 | 0.04 |
ENST00000588998.5
ENST00000586149.1 |
ECSIT
|
ECSIT signaling integrator |
| chr19_-_11529116 | 0.04 |
ENST00000592312.5
ENST00000590480.1 ENST00000585318.5 ENST00000270517.12 ENST00000252440.11 ENST00000417981.6 |
ECSIT
|
ECSIT signaling integrator |
| chr21_+_33403391 | 0.04 |
ENST00000290219.11
ENST00000381995.5 |
IFNGR2
|
interferon gamma receptor 2 |
| chr19_-_34773184 | 0.04 |
ENST00000588760.1
ENST00000329285.13 ENST00000587354.6 |
ZNF599
|
zinc finger protein 599 |
| chr2_+_218399838 | 0.03 |
ENST00000273062.7
|
CTDSP1
|
CTD small phosphatase 1 |
| chr1_-_211492111 | 0.03 |
ENST00000367002.5
ENST00000680073.1 |
RD3
|
RD3 regulator of GUCY2D |
| chr20_+_31968141 | 0.03 |
ENST00000562532.3
|
XKR7
|
XK related 7 |
| chr16_+_14974974 | 0.03 |
ENST00000535621.6
ENST00000396410.9 ENST00000566426.1 |
PDXDC1
|
pyridoxal dependent decarboxylase domain containing 1 |
| chr16_+_14975098 | 0.03 |
ENST00000569715.5
ENST00000627450.2 |
PDXDC1
|
pyridoxal dependent decarboxylase domain containing 1 |
| chr9_+_136665745 | 0.03 |
ENST00000371698.3
|
EGFL7
|
EGF like domain multiple 7 |
| chr1_-_224433776 | 0.03 |
ENST00000678879.1
ENST00000651911.2 |
WDR26
|
WD repeat domain 26 |
| chr14_-_105065422 | 0.02 |
ENST00000329797.8
ENST00000539291.6 ENST00000392585.2 |
GPR132
|
G protein-coupled receptor 132 |
| chr22_+_38468036 | 0.02 |
ENST00000409006.3
ENST00000216014.9 |
KDELR3
|
KDEL endoplasmic reticulum protein retention receptor 3 |
| chr10_+_102226293 | 0.02 |
ENST00000370005.4
|
ELOVL3
|
ELOVL fatty acid elongase 3 |
| chr6_-_32192845 | 0.02 |
ENST00000487761.5
|
GPSM3
|
G protein signaling modulator 3 |
| chr6_-_32192630 | 0.02 |
ENST00000375040.8
|
GPSM3
|
G protein signaling modulator 3 |
| chr8_+_93700524 | 0.02 |
ENST00000522324.5
ENST00000518322.6 ENST00000522803.5 ENST00000423990.6 ENST00000620645.1 |
CIBAR1
|
CBY1 interacting BAR domain containing 1 |
| chr17_+_44557476 | 0.02 |
ENST00000315323.5
|
FZD2
|
frizzled class receptor 2 |
| chr7_+_143381561 | 0.02 |
ENST00000354434.8
|
ZYX
|
zyxin |
| chr17_+_7884783 | 0.02 |
ENST00000380358.9
|
CHD3
|
chromodomain helicase DNA binding protein 3 |
| chr7_+_143381907 | 0.02 |
ENST00000392910.6
|
ZYX
|
zyxin |
| chr11_+_8019193 | 0.01 |
ENST00000534099.5
|
TUB
|
TUB bipartite transcription factor |
| chr1_+_218345326 | 0.01 |
ENST00000366930.9
|
TGFB2
|
transforming growth factor beta 2 |
| chrX_+_15790446 | 0.01 |
ENST00000380308.7
ENST00000307771.8 |
ZRSR2
|
zinc finger CCCH-type, RNA binding motif and serine/arginine rich 2 |
| chr15_-_66504832 | 0.01 |
ENST00000569438.2
ENST00000569696.5 ENST00000307961.11 |
RPL4
|
ribosomal protein L4 |
| chr12_+_15322480 | 0.01 |
ENST00000674188.1
ENST00000281171.9 ENST00000543886.6 |
PTPRO
|
protein tyrosine phosphatase receptor type O |
| chr5_+_145937793 | 0.01 |
ENST00000511217.1
|
SH3RF2
|
SH3 domain containing ring finger 2 |
| chrX_-_135296024 | 0.01 |
ENST00000370764.1
|
ZNF75D
|
zinc finger protein 75D |
| chr14_+_23555983 | 0.01 |
ENST00000404535.3
|
THTPA
|
thiamine triphosphatase |
| chr19_+_42220283 | 0.01 |
ENST00000301215.8
ENST00000597945.1 |
ZNF526
|
zinc finger protein 526 |
| chr22_-_21002081 | 0.01 |
ENST00000215742.9
ENST00000399133.2 |
THAP7
|
THAP domain containing 7 |
| chr9_+_17135017 | 0.00 |
ENST00000380641.4
ENST00000380647.8 |
CNTLN
|
centlein |
| chr2_+_203706475 | 0.00 |
ENST00000374481.7
ENST00000458610.6 |
CD28
|
CD28 molecule |
| chr19_+_40611863 | 0.00 |
ENST00000601032.5
|
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr1_+_6451304 | 0.00 |
ENST00000636644.1
|
ESPN
|
espin |
| chr6_+_26045374 | 0.00 |
ENST00000612966.3
|
H3C3
|
H3 clustered histone 3 |
| chr14_+_31561376 | 0.00 |
ENST00000550649.5
ENST00000281081.12 |
NUBPL
|
nucleotide binding protein like |
Network of associatons between targets according to the STRING database.
First level regulatory network of PRDM4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.0 | 0.1 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.0 | GO:0007538 | primary sex determination(GO:0007538) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990879 | CST complex(GO:1990879) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.0 | 0.0 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |