|
chr3_-_100993409
Show fit
|
1.75 |
ENST00000471714.6
|
ABI3BP
|
ABI family member 3 binding protein
|
|
chr3_-_100993507
Show fit
|
1.74 |
ENST00000284322.10
|
ABI3BP
|
ABI family member 3 binding protein
|
|
chr7_+_94394886
Show fit
|
1.14 |
ENST00000297268.11
ENST00000620463.1
|
COL1A2
|
collagen type I alpha 2 chain
|
|
chr14_-_52950992
Show fit
|
1.10 |
ENST00000343279.8
ENST00000399304.7
ENST00000395631.6
ENST00000341590.8
|
FERMT2
|
fermitin family member 2
|
|
chr10_+_31319125
Show fit
|
0.78 |
ENST00000320985.14
ENST00000560721.6
ENST00000558440.5
ENST00000424869.6
ENST00000542815.7
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr3_+_69763726
Show fit
|
0.63 |
ENST00000448226.9
|
MITF
|
melanocyte inducing transcription factor
|
|
chr2_-_144516154
Show fit
|
0.63 |
ENST00000637304.1
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr2_-_144431001
Show fit
|
0.62 |
ENST00000636413.1
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr2_-_144516397
Show fit
|
0.61 |
ENST00000638128.1
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr2_-_144430934
Show fit
|
0.61 |
ENST00000638087.1
ENST00000638007.1
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr2_-_79087986
Show fit
|
0.50 |
ENST00000305089.8
|
REG1B
|
regenerating family member 1 beta
|
|
chr12_+_65279092
Show fit
|
0.47 |
ENST00000646299.1
|
MSRB3
|
methionine sulfoxide reductase B3
|
|
chr10_+_122560639
Show fit
|
0.45 |
ENST00000344338.7
ENST00000330163.8
ENST00000652446.2
ENST00000666315.1
ENST00000368955.7
ENST00000368909.7
ENST00000368956.6
ENST00000619379.1
|
DMBT1
|
deleted in malignant brain tumors 1
|
|
chr1_-_85404494
Show fit
|
0.44 |
ENST00000633113.1
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr20_+_44582549
Show fit
|
0.44 |
ENST00000372886.6
|
PKIG
|
cAMP-dependent protein kinase inhibitor gamma
|
|
chr5_-_124744513
Show fit
|
0.44 |
ENST00000504926.5
|
ZNF608
|
zinc finger protein 608
|
|
chr12_+_65278919
Show fit
|
0.43 |
ENST00000538045.5
ENST00000642411.1
ENST00000535239.5
ENST00000614640.4
|
MSRB3
|
methionine sulfoxide reductase B3
|
|
chr10_+_122560679
Show fit
|
0.43 |
ENST00000657942.1
|
DMBT1
|
deleted in malignant brain tumors 1
|
|
chr10_+_122560751
Show fit
|
0.41 |
ENST00000338354.10
ENST00000664692.1
ENST00000653442.1
ENST00000664974.1
|
DMBT1
|
deleted in malignant brain tumors 1
|
|
chr9_-_101594995
Show fit
|
0.39 |
ENST00000636434.1
|
PPP3R2
|
protein phosphatase 3 regulatory subunit B, beta
|
|
chr10_-_37857582
Show fit
|
0.38 |
ENST00000395867.8
ENST00000611278.4
|
ZNF248
|
zinc finger protein 248
|
|
chr10_-_37858037
Show fit
|
0.35 |
ENST00000395873.7
ENST00000357328.8
ENST00000395874.2
|
ZNF248
|
zinc finger protein 248
|
|
chr3_+_69763549
Show fit
|
0.34 |
ENST00000472437.5
|
MITF
|
melanocyte inducing transcription factor
|
|
chr12_+_65279445
Show fit
|
0.33 |
ENST00000642404.1
|
MSRB3
|
methionine sulfoxide reductase B3
|
|
chr5_-_140564245
Show fit
|
0.30 |
ENST00000412920.7
ENST00000511201.2
ENST00000354402.9
ENST00000356738.6
|
APBB3
|
amyloid beta precursor protein binding family B member 3
|
|
chr5_-_88823763
Show fit
|
0.29 |
ENST00000635898.1
ENST00000626391.2
ENST00000628656.2
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr5_-_88877967
Show fit
|
0.28 |
ENST00000508610.5
ENST00000636294.1
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr2_+_109898685
Show fit
|
0.28 |
ENST00000480744.2
|
LIMS3
|
LIM zinc finger domain containing 3
|
|
chr17_+_2796404
Show fit
|
0.27 |
ENST00000366401.8
ENST00000254695.13
ENST00000542807.1
|
RAP1GAP2
|
RAP1 GTPase activating protein 2
|
|
chr12_+_6828377
Show fit
|
0.26 |
ENST00000290510.10
|
P3H3
|
prolyl 3-hydroxylase 3
|
|
chr10_-_33335074
Show fit
|
0.26 |
ENST00000432372.6
|
NRP1
|
neuropilin 1
|
|
chr12_+_12785652
Show fit
|
0.25 |
ENST00000356591.5
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr13_-_74133892
Show fit
|
0.25 |
ENST00000377669.7
|
KLF12
|
Kruppel like factor 12
|
|
chr10_-_33334898
Show fit
|
0.25 |
ENST00000395995.5
|
NRP1
|
neuropilin 1
|
|
chr10_-_33334625
Show fit
|
0.23 |
ENST00000374875.5
ENST00000374822.8
ENST00000374867.7
|
NRP1
|
neuropilin 1
|
|
chr7_-_120858066
Show fit
|
0.22 |
ENST00000222747.8
|
TSPAN12
|
tetraspanin 12
|
|
chr7_+_90709816
Show fit
|
0.22 |
ENST00000436577.3
|
CDK14
|
cyclin dependent kinase 14
|
|
chr5_+_141412979
Show fit
|
0.22 |
ENST00000612503.1
ENST00000398610.3
|
PCDHGA10
|
protocadherin gamma subfamily A, 10
|
|
chr7_+_90709530
Show fit
|
0.22 |
ENST00000406263.5
|
CDK14
|
cyclin dependent kinase 14
|
|
chr9_-_137028878
Show fit
|
0.21 |
ENST00000625103.1
ENST00000614293.4
|
ABCA2
|
ATP binding cassette subfamily A member 2
|
|
chr19_-_39204254
Show fit
|
0.21 |
ENST00000318438.7
|
SYCN
|
syncollin
|
|
chr14_-_52791597
Show fit
|
0.21 |
ENST00000216410.8
ENST00000557604.1
|
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1
|
|
chr11_+_118607598
Show fit
|
0.20 |
ENST00000600882.6
ENST00000356063.9
|
PHLDB1
|
pleckstrin homology like domain family B member 1
|
|
chr10_-_43648845
Show fit
|
0.20 |
ENST00000374433.7
|
ZNF32
|
zinc finger protein 32
|
|
chr10_-_43648704
Show fit
|
0.20 |
ENST00000395797.1
|
ZNF32
|
zinc finger protein 32
|
|
chr19_-_55597753
Show fit
|
0.20 |
ENST00000587678.1
|
FIZ1
|
FLT3 interacting zinc finger 1
|
|
chr10_-_33334382
Show fit
|
0.19 |
ENST00000374823.9
ENST00000374821.9
ENST00000374816.7
|
NRP1
|
neuropilin 1
|
|
chr2_-_110473041
Show fit
|
0.19 |
ENST00000632897.1
ENST00000413601.3
|
LIMS4
|
LIM zinc finger domain containing 4
|
|
chr7_+_90709231
Show fit
|
0.19 |
ENST00000446790.5
ENST00000265741.7
|
CDK14
|
cyclin dependent kinase 14
|
|
chr21_+_37420299
Show fit
|
0.18 |
ENST00000455097.6
ENST00000643854.1
ENST00000645424.1
ENST00000642309.1
ENST00000645774.1
ENST00000398956.2
|
DYRK1A
|
dual specificity tyrosine phosphorylation regulated kinase 1A
|
|
chr15_+_40405787
Show fit
|
0.18 |
ENST00000610693.5
ENST00000479013.7
ENST00000487418.8
|
IVD
|
isovaleryl-CoA dehydrogenase
|
|
chr14_+_51489112
Show fit
|
0.17 |
ENST00000356218.8
|
FRMD6
|
FERM domain containing 6
|
|
chr4_+_26319636
Show fit
|
0.17 |
ENST00000342295.6
ENST00000506956.5
ENST00000512671.6
ENST00000345843.8
|
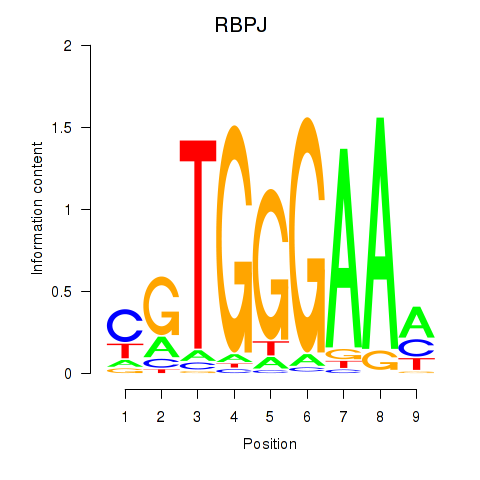
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region
|
|
chr1_+_61404076
Show fit
|
0.16 |
ENST00000357977.5
|
NFIA
|
nuclear factor I A
|
|
chr10_+_113679839
Show fit
|
0.16 |
ENST00000369318.8
ENST00000369315.5
|
CASP7
|
caspase 7
|
|
chr11_+_118607579
Show fit
|
0.16 |
ENST00000530708.4
|
PHLDB1
|
pleckstrin homology like domain family B member 1
|
|
chrX_+_106802660
Show fit
|
0.16 |
ENST00000357242.10
ENST00000310452.6
ENST00000481617.6
ENST00000276175.7
|
TBC1D8B
|
TBC1 domain family member 8B
|
|
chr9_-_101594918
Show fit
|
0.16 |
ENST00000374806.2
|
PPP3R2
|
protein phosphatase 3 regulatory subunit B, beta
|
|
chr10_+_113679523
Show fit
|
0.15 |
ENST00000345633.8
ENST00000614447.4
ENST00000369321.6
|
CASP7
|
caspase 7
|
|
chr11_+_64285533
Show fit
|
0.15 |
ENST00000538032.5
|
GPR137
|
G protein-coupled receptor 137
|
|
chr19_-_50025936
Show fit
|
0.15 |
ENST00000596445.5
ENST00000599538.5
|
VRK3
|
VRK serine/threonine kinase 3
|
|
chr6_+_33208488
Show fit
|
0.15 |
ENST00000374656.5
|
RING1
|
ring finger protein 1
|
|
chr2_-_40430257
Show fit
|
0.14 |
ENST00000408028.6
ENST00000332839.8
ENST00000406391.2
ENST00000405901.7
|
SLC8A1
|
solute carrier family 8 member A1
|
|
chr11_-_119340544
Show fit
|
0.14 |
ENST00000530681.2
|
C1QTNF5
|
C1q and TNF related 5
|
|
chr12_-_89656093
Show fit
|
0.14 |
ENST00000359142.7
|
ATP2B1
|
ATPase plasma membrane Ca2+ transporting 1
|
|
chr12_-_89656051
Show fit
|
0.13 |
ENST00000261173.6
|
ATP2B1
|
ATPase plasma membrane Ca2+ transporting 1
|
|
chr10_-_27242068
Show fit
|
0.13 |
ENST00000375901.5
ENST00000412279.1
ENST00000676731.1
ENST00000679220.1
ENST00000678392.1
ENST00000678446.1
ENST00000677441.1
ENST00000375905.8
|
ACBD5
|
acyl-CoA binding domain containing 5
|
|
chr12_+_57772587
Show fit
|
0.12 |
ENST00000300209.13
|
EEF1AKMT3
|
EEF1A lysine methyltransferase 3
|
|
chr3_-_100993448
Show fit
|
0.12 |
ENST00000495063.6
ENST00000486770.7
ENST00000530539.2
|
ABI3BP
|
ABI family member 3 binding protein
|
|
chr16_+_23302292
Show fit
|
0.12 |
ENST00000343070.7
|
SCNN1B
|
sodium channel epithelial 1 subunit beta
|
|
chr17_-_41481140
Show fit
|
0.12 |
ENST00000246639.6
ENST00000393989.1
|
KRT35
|
keratin 35
|
|
chr17_-_38735577
Show fit
|
0.12 |
ENST00000610747.1
|
PCGF2
|
polycomb group ring finger 2
|
|
chr10_+_73168111
Show fit
|
0.11 |
ENST00000242505.11
|
FAM149B1
|
family with sequence similarity 149 member B1
|
|
chr1_-_180502536
Show fit
|
0.11 |
ENST00000367595.4
|
ACBD6
|
acyl-CoA binding domain containing 6
|
|
chr11_-_124935973
Show fit
|
0.11 |
ENST00000298251.5
|
HEPACAM
|
hepatic and glial cell adhesion molecule
|
|
chr5_+_138179145
Show fit
|
0.11 |
ENST00000508792.5
ENST00000504621.1
|
KIF20A
|
kinesin family member 20A
|
|
chr22_-_31489761
Show fit
|
0.11 |
ENST00000344710.9
ENST00000330125.10
ENST00000397518.5
|
EIF4ENIF1
|
eukaryotic translation initiation factor 4E nuclear import factor 1
|
|
chr1_+_36224410
Show fit
|
0.11 |
ENST00000469141.6
ENST00000648638.1
ENST00000354618.10
|
THRAP3
|
thyroid hormone receptor associated protein 3
|
|
chr8_+_67952028
Show fit
|
0.11 |
ENST00000288368.5
|
PREX2
|
phosphatidylinositol-3,4,5-trisphosphate dependent Rac exchange factor 2
|
|
chr7_+_74209947
Show fit
|
0.11 |
ENST00000475494.5
ENST00000398475.5
|
LAT2
|
linker for activation of T cells family member 2
|
|
chr10_+_111077021
Show fit
|
0.11 |
ENST00000280155.4
|
ADRA2A
|
adrenoceptor alpha 2A
|
|
chr1_-_197775435
Show fit
|
0.10 |
ENST00000620048.6
|
DENND1B
|
DENN domain containing 1B
|
|
chr1_-_180502811
Show fit
|
0.10 |
ENST00000642319.1
|
ACBD6
|
acyl-CoA binding domain containing 6
|
|
chr12_+_4809176
Show fit
|
0.10 |
ENST00000280684.3
|
KCNA6
|
potassium voltage-gated channel subfamily A member 6
|
|
chr12_+_57772648
Show fit
|
0.10 |
ENST00000333012.5
|
EEF1AKMT3
|
EEF1A lysine methyltransferase 3
|
|
chr11_+_64285472
Show fit
|
0.10 |
ENST00000535675.5
ENST00000543383.5
|
GPR137
|
G protein-coupled receptor 137
|
|
chr7_-_27130182
Show fit
|
0.10 |
ENST00000511914.1
|
HOXA4
|
homeobox A4
|
|
chr3_-_45915698
Show fit
|
0.10 |
ENST00000539217.5
|
LZTFL1
|
leucine zipper transcription factor like 1
|
|
chr5_+_138179093
Show fit
|
0.10 |
ENST00000394894.8
|
KIF20A
|
kinesin family member 20A
|
|
chr5_+_42423433
Show fit
|
0.09 |
ENST00000230882.9
|
GHR
|
growth hormone receptor
|
|
chr10_-_48251757
Show fit
|
0.09 |
ENST00000305531.3
|
FRMPD2
|
FERM and PDZ domain containing 2
|
|
chr1_-_43389768
Show fit
|
0.09 |
ENST00000372455.4
ENST00000372457.9
ENST00000290663.10
|
MED8
|
mediator complex subunit 8
|
|
chrX_+_38006551
Show fit
|
0.09 |
ENST00000297875.7
|
SYTL5
|
synaptotagmin like 5
|
|
chr1_+_113390495
Show fit
|
0.09 |
ENST00000307546.14
|
MAGI3
|
membrane associated guanylate kinase, WW and PDZ domain containing 3
|
|
chr18_+_3451585
Show fit
|
0.09 |
ENST00000551541.5
|
TGIF1
|
TGFB induced factor homeobox 1
|
|
chr14_-_24232332
Show fit
|
0.09 |
ENST00000534348.5
ENST00000524927.1
ENST00000250495.10
|
NEDD8-MDP1
NEDD8
|
NEDD8-MDP1 readthrough
NEDD8 ubiquitin like modifier
|
|
chr12_-_4649043
Show fit
|
0.09 |
ENST00000545990.6
ENST00000228850.6
|
AKAP3
|
A-kinase anchoring protein 3
|
|
chr17_-_28726186
Show fit
|
0.08 |
ENST00000292090.8
|
TLCD1
|
TLC domain containing 1
|
|
chr19_-_38899529
Show fit
|
0.08 |
ENST00000249396.12
ENST00000437828.5
|
SIRT2
|
sirtuin 2
|
|
chr3_+_159852933
Show fit
|
0.08 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr1_+_151008412
Show fit
|
0.08 |
ENST00000271620.8
ENST00000650332.1
ENST00000450884.5
ENST00000368937.5
ENST00000431193.5
ENST00000368936.5
|
PRUNE1
|
prune exopolyphosphatase 1
|
|
chr15_+_67125707
Show fit
|
0.08 |
ENST00000540846.6
|
SMAD3
|
SMAD family member 3
|
|
chr1_-_151008365
Show fit
|
0.08 |
ENST00000361936.9
ENST00000361738.11
|
MINDY1
|
MINDY lysine 48 deubiquitinase 1
|
|
chr17_-_16215520
Show fit
|
0.08 |
ENST00000582357.5
ENST00000436828.5
ENST00000268712.8
ENST00000411510.5
ENST00000395857.7
|
NCOR1
|
nuclear receptor corepressor 1
|
|
chr8_-_22232020
Show fit
|
0.08 |
ENST00000454243.7
ENST00000321613.7
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein
|
|
chr4_+_26320975
Show fit
|
0.08 |
ENST00000509158.6
ENST00000681856.1
ENST00000680140.1
ENST00000355476.8
ENST00000680511.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region
|
|
chr19_-_38899800
Show fit
|
0.08 |
ENST00000414941.5
ENST00000358931.9
ENST00000392081.6
|
SIRT2
|
sirtuin 2
|
|
chr19_+_3094348
Show fit
|
0.08 |
ENST00000078429.9
|
GNA11
|
G protein subunit alpha 11
|
|
chr3_-_19934189
Show fit
|
0.07 |
ENST00000295824.14
|
EFHB
|
EF-hand domain family member B
|
|
chr16_-_70685791
Show fit
|
0.07 |
ENST00000616026.4
|
MTSS2
|
MTSS I-BAR domain containing 2
|
|
chr19_+_36687579
Show fit
|
0.07 |
ENST00000682579.1
ENST00000536254.6
|
ZNF567
|
zinc finger protein 567
|
|
chr1_+_151008513
Show fit
|
0.07 |
ENST00000368935.1
|
PRUNE1
|
prune exopolyphosphatase 1
|
|
chr16_-_1611985
Show fit
|
0.07 |
ENST00000426508.7
|
IFT140
|
intraflagellar transport 140
|
|
chr19_-_45584769
Show fit
|
0.07 |
ENST00000263275.5
|
OPA3
|
outer mitochondrial membrane lipid metabolism regulator OPA3
|
|
chr9_+_70043840
Show fit
|
0.07 |
ENST00000377182.5
|
MAMDC2
|
MAM domain containing 2
|
|
chr6_+_20403679
Show fit
|
0.07 |
ENST00000535432.2
|
E2F3
|
E2F transcription factor 3
|
|
chr1_-_999981
Show fit
|
0.06 |
ENST00000484667.2
|
HES4
|
hes family bHLH transcription factor 4
|
|
chr20_+_33662310
Show fit
|
0.06 |
ENST00000375222.4
|
C20orf144
|
chromosome 20 open reading frame 144
|
|
chr19_-_40218339
Show fit
|
0.06 |
ENST00000311308.6
|
TTC9B
|
tetratricopeptide repeat domain 9B
|
|
chr19_+_43596480
Show fit
|
0.06 |
ENST00000533118.5
|
ZNF576
|
zinc finger protein 576
|
|
chr16_+_85611401
Show fit
|
0.06 |
ENST00000405402.6
|
GSE1
|
Gse1 coiled-coil protein
|
|
chr11_+_64285219
Show fit
|
0.06 |
ENST00000377702.8
|
GPR137
|
G protein-coupled receptor 137
|
|
chr11_-_64744811
Show fit
|
0.06 |
ENST00000377497.7
ENST00000377487.5
ENST00000430645.1
|
RASGRP2
|
RAS guanyl releasing protein 2
|
|
chr11_-_31817904
Show fit
|
0.06 |
ENST00000423822.7
|
PAX6
|
paired box 6
|
|
chr11_-_31817937
Show fit
|
0.06 |
ENST00000638965.1
ENST00000241001.13
ENST00000638914.3
ENST00000379132.8
ENST00000379129.7
|
PAX6
|
paired box 6
|
|
chr13_-_19782970
Show fit
|
0.06 |
ENST00000427943.1
ENST00000619300.4
|
PSPC1
|
paraspeckle component 1
|
|
chr16_+_53703963
Show fit
|
0.06 |
ENST00000636218.1
ENST00000637001.1
ENST00000471389.6
ENST00000637969.1
ENST00000640179.1
|
FTO
|
FTO alpha-ketoglutarate dependent dioxygenase
|
|
chr15_+_74890005
Show fit
|
0.06 |
ENST00000569931.5
ENST00000566377.5
ENST00000569233.5
ENST00000567132.5
ENST00000564633.5
ENST00000568907.5
ENST00000563422.5
ENST00000564003.5
ENST00000562800.5
ENST00000563786.5
ENST00000535694.5
ENST00000323744.10
ENST00000352410.9
ENST00000568828.5
ENST00000562606.5
ENST00000565576.5
ENST00000567570.5
|
MPI
|
mannose phosphate isomerase
|
|
chr7_+_100589573
Show fit
|
0.05 |
ENST00000468962.5
ENST00000427939.2
|
FBXO24
|
F-box protein 24
|
|
chr3_-_184017863
Show fit
|
0.05 |
ENST00000427120.6
ENST00000334444.11
ENST00000392579.6
ENST00000382494.6
ENST00000265586.10
ENST00000446941.2
|
ABCC5
|
ATP binding cassette subfamily C member 5
|
|
chr7_-_120857124
Show fit
|
0.05 |
ENST00000441017.5
ENST00000424710.5
ENST00000433758.5
|
TSPAN12
|
tetraspanin 12
|
|
chr8_-_100336184
Show fit
|
0.05 |
ENST00000519527.5
ENST00000522369.5
|
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase
|
|
chrX_-_70260199
Show fit
|
0.05 |
ENST00000374519.4
|
P2RY4
|
pyrimidinergic receptor P2Y4
|
|
chr16_+_75148478
Show fit
|
0.05 |
ENST00000568079.5
ENST00000570010.6
ENST00000464850.5
ENST00000332307.4
ENST00000393430.6
|
ZFP1
|
ZFP1 zinc finger protein
|
|
chr9_-_95507416
Show fit
|
0.05 |
ENST00000429896.6
|
PTCH1
|
patched 1
|
|
chr16_+_58025745
Show fit
|
0.04 |
ENST00000219271.4
|
MMP15
|
matrix metallopeptidase 15
|
|
chr19_-_45584810
Show fit
|
0.04 |
ENST00000323060.3
|
OPA3
|
outer mitochondrial membrane lipid metabolism regulator OPA3
|
|
chr16_+_30064142
Show fit
|
0.04 |
ENST00000562168.5
ENST00000569545.5
|
ALDOA
|
aldolase, fructose-bisphosphate A
|
|
chr20_+_31637905
Show fit
|
0.04 |
ENST00000376075.4
|
COX4I2
|
cytochrome c oxidase subunit 4I2
|
|
chr8_-_67062120
Show fit
|
0.04 |
ENST00000357849.9
|
COPS5
|
COP9 signalosome subunit 5
|
|
chr13_-_19782923
Show fit
|
0.04 |
ENST00000338910.9
|
PSPC1
|
paraspeckle component 1
|
|
chr1_+_7961894
Show fit
|
0.04 |
ENST00000377491.5
ENST00000377488.5
|
PARK7
|
Parkinsonism associated deglycase
|
|
chr20_-_46308485
Show fit
|
0.04 |
ENST00000537909.4
|
CDH22
|
cadherin 22
|
|
chr17_+_43170452
Show fit
|
0.04 |
ENST00000341165.10
ENST00000586650.5
|
NBR1
|
NBR1 autophagy cargo receptor
|
|
chr4_+_26320782
Show fit
|
0.04 |
ENST00000514807.5
ENST00000348160.9
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region
|
|
chr1_-_91021996
Show fit
|
0.04 |
ENST00000337393.10
|
ZNF644
|
zinc finger protein 644
|
|
chr4_+_26320563
Show fit
|
0.04 |
ENST00000361572.10
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region
|
|
chr19_+_34926892
Show fit
|
0.04 |
ENST00000303586.11
ENST00000601142.2
ENST00000439785.5
ENST00000601540.5
ENST00000601957.5
|
ZNF30
|
zinc finger protein 30
|
|
chr16_+_30064274
Show fit
|
0.04 |
ENST00000563060.6
|
ALDOA
|
aldolase, fructose-bisphosphate A
|
|
chr12_+_10505602
Show fit
|
0.04 |
ENST00000322446.3
|
EIF2S3B
|
eukaryotic translation initiation factor 2 subunit gamma B
|
|
chr4_+_70195719
Show fit
|
0.04 |
ENST00000683306.1
|
ODAM
|
odontogenic, ameloblast associated
|
|
chr6_-_75493773
Show fit
|
0.03 |
ENST00000237172.12
|
FILIP1
|
filamin A interacting protein 1
|
|
chr5_-_138178599
Show fit
|
0.03 |
ENST00000454473.5
ENST00000418329.5
ENST00000254900.10
ENST00000230901.9
ENST00000402931.5
ENST00000411594.6
ENST00000430331.1
|
BRD8
|
bromodomain containing 8
|
|
chr12_-_31792290
Show fit
|
0.03 |
ENST00000340398.5
|
H3-5
|
H3.5 histone
|
|
chr1_-_197775298
Show fit
|
0.03 |
ENST00000367396.7
|
DENND1B
|
DENN domain containing 1B
|
|
chr1_-_147172456
Show fit
|
0.03 |
ENST00000254101.4
|
PRKAB2
|
protein kinase AMP-activated non-catalytic subunit beta 2
|
|
chr11_-_64879675
Show fit
|
0.03 |
ENST00000359393.6
ENST00000433803.1
ENST00000411683.1
|
EHD1
|
EH domain containing 1
|
|
chr1_+_43389889
Show fit
|
0.03 |
ENST00000562955.2
ENST00000634258.3
|
SZT2
|
SZT2 subunit of KICSTOR complex
|
|
chr1_-_204166334
Show fit
|
0.03 |
ENST00000272190.9
|
REN
|
renin
|
|
chr1_+_50106265
Show fit
|
0.03 |
ENST00000357083.8
|
ELAVL4
|
ELAV like RNA binding protein 4
|
|
chr7_+_30771388
Show fit
|
0.03 |
ENST00000265299.6
|
MINDY4
|
MINDY lysine 48 deubiquitinase 4
|
|
chr17_-_64662290
Show fit
|
0.03 |
ENST00000262435.14
|
SMURF2
|
SMAD specific E3 ubiquitin protein ligase 2
|
|
chr2_+_186486246
Show fit
|
0.03 |
ENST00000337859.11
|
ZC3H15
|
zinc finger CCCH-type containing 15
|
|
chr18_+_13218195
Show fit
|
0.03 |
ENST00000679167.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4
|
|
chr11_-_119340816
Show fit
|
0.03 |
ENST00000528368.3
|
C1QTNF5
|
C1q and TNF related 5
|
|
chr11_+_64285839
Show fit
|
0.03 |
ENST00000438980.7
ENST00000540370.1
|
GPR137
|
G protein-coupled receptor 137
|
|
chr3_+_156142962
Show fit
|
0.03 |
ENST00000471742.5
|
KCNAB1
|
potassium voltage-gated channel subfamily A member regulatory beta subunit 1
|
|
chr2_+_149330506
Show fit
|
0.03 |
ENST00000334166.9
|
LYPD6
|
LY6/PLAUR domain containing 6
|
|
chr6_-_26056460
Show fit
|
0.03 |
ENST00000343677.4
|
H1-2
|
H1.2 linker histone, cluster member
|
|
chr7_+_102912983
Show fit
|
0.02 |
ENST00000339431.9
ENST00000249377.4
|
LRRC17
|
leucine rich repeat containing 17
|
|
chr11_-_64879709
Show fit
|
0.02 |
ENST00000621096.4
|
EHD1
|
EH domain containing 1
|
|
chr19_+_39997031
Show fit
|
0.02 |
ENST00000599504.5
ENST00000596894.5
ENST00000601138.5
ENST00000347077.9
ENST00000600094.5
|
ZNF546
|
zinc finger protein 546
|
|
chr1_+_43389874
Show fit
|
0.02 |
ENST00000372450.8
|
SZT2
|
SZT2 subunit of KICSTOR complex
|
|
chr11_-_2270584
Show fit
|
0.02 |
ENST00000331289.5
|
ASCL2
|
achaete-scute family bHLH transcription factor 2
|
|
chr1_+_109483905
Show fit
|
0.02 |
ENST00000459635.2
ENST00000683729.1
ENST00000369870.7
|
ATXN7L2
|
ataxin 7 like 2
|
|
chr14_+_24232422
Show fit
|
0.02 |
ENST00000620807.4
ENST00000355299.8
ENST00000559836.5
|
GMPR2
|
guanosine monophosphate reductase 2
|
|
chr2_+_169733811
Show fit
|
0.02 |
ENST00000392647.7
|
KLHL23
|
kelch like family member 23
|
|
chr10_-_73167961
Show fit
|
0.02 |
ENST00000372979.9
|
ECD
|
ecdysoneless cell cycle regulator
|
|
chr19_+_43596388
Show fit
|
0.02 |
ENST00000391965.6
ENST00000525771.1
|
ZNF576
|
zinc finger protein 576
|
|
chr15_+_67065586
Show fit
|
0.02 |
ENST00000327367.9
|
SMAD3
|
SMAD family member 3
|
|
chr11_-_67523396
Show fit
|
0.02 |
ENST00000353903.9
ENST00000294288.5
|
CABP2
|
calcium binding protein 2
|
|
chr6_-_32224060
Show fit
|
0.02 |
ENST00000375023.3
|
NOTCH4
|
notch receptor 4
|
|
chr4_+_155667654
Show fit
|
0.02 |
ENST00000513574.1
|
GUCY1A1
|
guanylate cyclase 1 soluble subunit alpha 1
|
|
chr7_+_128862841
Show fit
|
0.02 |
ENST00000249289.5
ENST00000492758.1
|
ATP6V1F
|
ATPase H+ transporting V1 subunit F
|
|
chr10_-_73168052
Show fit
|
0.02 |
ENST00000430082.6
ENST00000454759.6
ENST00000413026.1
ENST00000453402.5
|
ECD
|
ecdysoneless cell cycle regulator
|
|
chr10_+_116545907
Show fit
|
0.02 |
ENST00000369221.2
|
PNLIP
|
pancreatic lipase
|
|
chr19_+_19405655
Show fit
|
0.01 |
ENST00000683918.1
|
GATAD2A
|
GATA zinc finger domain containing 2A
|
|
chrX_-_49264668
Show fit
|
0.01 |
ENST00000455775.7
ENST00000652559.1
ENST00000376207.10
ENST00000557224.6
ENST00000684155.1
ENST00000376199.7
|
FOXP3
|
forkhead box P3
|
|
chr7_-_151632330
Show fit
|
0.01 |
ENST00000418337.6
|
PRKAG2
|
protein kinase AMP-activated non-catalytic subunit gamma 2
|
|
chr11_-_64745331
Show fit
|
0.01 |
ENST00000377489.5
ENST00000354024.7
|
RASGRP2
|
RAS guanyl releasing protein 2
|
|
chr11_-_32430811
Show fit
|
0.01 |
ENST00000379079.8
ENST00000530998.5
|
WT1
|
WT1 transcription factor
|
|
chr2_+_218959635
Show fit
|
0.01 |
ENST00000302625.6
|
CDK5R2
|
cyclin dependent kinase 5 regulatory subunit 2
|
|
chr15_-_52529050
Show fit
|
0.01 |
ENST00000399231.7
|
MYO5A
|
myosin VA
|
|
chr16_-_53703883
Show fit
|
0.01 |
ENST00000262135.9
ENST00000564374.5
ENST00000566096.5
|
RPGRIP1L
|
RPGRIP1 like
|
|
chr3_-_132037800
Show fit
|
0.01 |
ENST00000617767.4
|
CPNE4
|
copine 4
|
|
chr1_-_23369813
Show fit
|
0.01 |
ENST00000314011.9
|
ZNF436
|
zinc finger protein 436
|
|
chr13_+_39038347
Show fit
|
0.01 |
ENST00000379599.6
ENST00000379600.8
|
NHLRC3
|
NHL repeat containing 3
|
|
chr10_-_97445850
Show fit
|
0.01 |
ENST00000477692.6
ENST00000485122.6
ENST00000370886.9
ENST00000370885.8
ENST00000370902.8
ENST00000370884.5
|
EXOSC1
|
exosome component 1
|
|
chr12_-_7695752
Show fit
|
0.01 |
ENST00000329913.4
|
GDF3
|
growth differentiation factor 3
|
|
chr21_-_6499202
Show fit
|
0.01 |
ENST00000619610.2
ENST00000610664.5
ENST00000639996.1
|
U2AF1L5
|
U2 small nuclear RNA auxiliary factor 1 like 5
|