|
chr18_+_23949847
Show fit
|
2.42 |
ENST00000588004.1
|
LAMA3
|
laminin subunit alpha 3
|
|
chr18_+_23873000
Show fit
|
1.92 |
ENST00000269217.11
ENST00000587184.5
|
LAMA3
|
laminin subunit alpha 3
|
|
chr6_+_137867241
Show fit
|
1.38 |
ENST00000612899.5
ENST00000420009.5
|
TNFAIP3
|
TNF alpha induced protein 3
|
|
chr6_+_137867414
Show fit
|
1.37 |
ENST00000237289.8
ENST00000433680.1
|
TNFAIP3
|
TNF alpha induced protein 3
|
|
chr12_-_57767057
Show fit
|
1.11 |
ENST00000228606.9
|
CYP27B1
|
cytochrome P450 family 27 subfamily B member 1
|
|
chr18_+_36297661
Show fit
|
1.09 |
ENST00000257209.8
ENST00000590592.5
ENST00000359247.8
|
FHOD3
|
formin homology 2 domain containing 3
|
|
chr10_-_98268186
Show fit
|
0.99 |
ENST00000260702.4
|
LOXL4
|
lysyl oxidase like 4
|
|
chr12_-_66678934
Show fit
|
0.81 |
ENST00000545666.5
ENST00000398016.7
ENST00000359742.9
ENST00000538211.5
|
GRIP1
|
glutamate receptor interacting protein 1
|
|
chr1_-_209651291
Show fit
|
0.78 |
ENST00000391911.5
ENST00000415782.1
|
LAMB3
|
laminin subunit beta 3
|
|
chr1_+_200894892
Show fit
|
0.76 |
ENST00000413687.3
|
INAVA
|
innate immunity activator
|
|
chr20_+_45174894
Show fit
|
0.73 |
ENST00000243924.4
|
PI3
|
peptidase inhibitor 3
|
|
chr17_-_58415628
Show fit
|
0.65 |
ENST00000583753.5
|
RNF43
|
ring finger protein 43
|
|
chr19_-_4338786
Show fit
|
0.65 |
ENST00000601482.1
ENST00000600324.5
ENST00000594605.6
|
STAP2
|
signal transducing adaptor family member 2
|
|
chr11_+_10455292
Show fit
|
0.63 |
ENST00000396553.6
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr11_+_102317492
Show fit
|
0.63 |
ENST00000673846.1
|
BIRC3
|
baculoviral IAP repeat containing 3
|
|
chr6_-_143511682
Show fit
|
0.62 |
ENST00000002165.11
|
FUCA2
|
alpha-L-fucosidase 2
|
|
chr11_-_58578096
Show fit
|
0.61 |
ENST00000528954.5
ENST00000528489.1
|
LPXN
|
leupaxin
|
|
chr4_+_73869385
Show fit
|
0.59 |
ENST00000395761.4
|
CXCL1
|
C-X-C motif chemokine ligand 1
|
|
chr6_+_30882914
Show fit
|
0.56 |
ENST00000509639.5
ENST00000412274.6
ENST00000507901.5
ENST00000507046.5
ENST00000437124.6
ENST00000454612.6
ENST00000396342.6
|
DDR1
|
discoidin domain receptor tyrosine kinase 1
|
|
chr12_-_94616061
Show fit
|
0.54 |
ENST00000551457.1
|
TMCC3
|
transmembrane and coiled-coil domain family 3
|
|
chr1_+_95117324
Show fit
|
0.53 |
ENST00000370203.9
ENST00000456991.5
|
TLCD4
|
TLC domain containing 4
|
|
chr12_-_85836372
Show fit
|
0.52 |
ENST00000361228.5
|
RASSF9
|
Ras association domain family member 9
|
|
chr1_-_9129085
Show fit
|
0.49 |
ENST00000377411.5
|
GPR157
|
G protein-coupled receptor 157
|
|
chr19_-_50968775
Show fit
|
0.47 |
ENST00000391808.5
|
KLK6
|
kallikrein related peptidase 6
|
|
chr2_+_162318884
Show fit
|
0.46 |
ENST00000446271.5
ENST00000429691.6
|
GCA
|
grancalcin
|
|
chr19_-_50968125
Show fit
|
0.43 |
ENST00000594641.1
|
KLK6
|
kallikrein related peptidase 6
|
|
chr6_+_13924867
Show fit
|
0.41 |
ENST00000544682.5
ENST00000420478.2
|
RNF182
|
ring finger protein 182
|
|
chr16_+_27313879
Show fit
|
0.41 |
ENST00000562142.5
ENST00000561742.5
ENST00000543915.6
ENST00000395762.7
ENST00000563002.5
|
IL4R
|
interleukin 4 receptor
|
|
chr12_+_51238854
Show fit
|
0.39 |
ENST00000549732.6
ENST00000604900.5
|
DAZAP2
|
DAZ associated protein 2
|
|
chr7_-_16804987
Show fit
|
0.39 |
ENST00000401412.5
ENST00000419304.7
|
AGR2
|
anterior gradient 2, protein disulphide isomerase family member
|
|
chr11_-_441964
Show fit
|
0.39 |
ENST00000332826.7
|
ANO9
|
anoctamin 9
|
|
chr12_-_122500520
Show fit
|
0.37 |
ENST00000540586.1
ENST00000543897.5
|
ZCCHC8
|
zinc finger CCHC-type containing 8
|
|
chr2_-_162318475
Show fit
|
0.36 |
ENST00000648433.1
|
IFIH1
|
interferon induced with helicase C domain 1
|
|
chr19_+_14031746
Show fit
|
0.36 |
ENST00000263379.4
|
IL27RA
|
interleukin 27 receptor subunit alpha
|
|
chr1_-_204411804
Show fit
|
0.36 |
ENST00000367188.5
|
PPP1R15B
|
protein phosphatase 1 regulatory subunit 15B
|
|
chr9_+_128149447
Show fit
|
0.36 |
ENST00000277480.7
ENST00000372998.1
|
LCN2
|
lipocalin 2
|
|
chr14_+_23372809
Show fit
|
0.35 |
ENST00000397242.2
ENST00000329715.2
|
IL25
|
interleukin 25
|
|
chr2_+_219229783
Show fit
|
0.35 |
ENST00000453432.5
ENST00000409849.5
ENST00000323348.10
ENST00000416565.1
ENST00000410034.7
ENST00000447157.5
|
ANKZF1
|
ankyrin repeat and zinc finger peptidyl tRNA hydrolase 1
|
|
chr14_-_24188787
Show fit
|
0.34 |
ENST00000625289.1
ENST00000354464.11
|
IPO4
|
importin 4
|
|
chr7_-_82443715
Show fit
|
0.34 |
ENST00000356253.9
ENST00000423588.1
|
CACNA2D1
|
calcium voltage-gated channel auxiliary subunit alpha2delta 1
|
|
chr11_-_72781833
Show fit
|
0.32 |
ENST00000535054.1
ENST00000545082.5
|
STARD10
|
StAR related lipid transfer domain containing 10
|
|
chr11_-_72781858
Show fit
|
0.32 |
ENST00000537947.5
|
STARD10
|
StAR related lipid transfer domain containing 10
|
|
chr6_+_14117764
Show fit
|
0.32 |
ENST00000379153.4
|
CD83
|
CD83 molecule
|
|
chr12_-_54981838
Show fit
|
0.31 |
ENST00000316577.12
|
TESPA1
|
thymocyte expressed, positive selection associated 1
|
|
chr9_-_133129395
Show fit
|
0.30 |
ENST00000393157.8
|
RALGDS
|
ral guanine nucleotide dissociation stimulator
|
|
chr10_+_102395693
Show fit
|
0.29 |
ENST00000652277.1
ENST00000189444.11
ENST00000661543.1
|
NFKB2
|
nuclear factor kappa B subunit 2
|
|
chr4_-_74099187
Show fit
|
0.29 |
ENST00000508487.3
|
CXCL2
|
C-X-C motif chemokine ligand 2
|
|
chr6_+_32154131
Show fit
|
0.29 |
ENST00000375143.6
ENST00000324816.11
ENST00000424499.1
|
PPT2
|
palmitoyl-protein thioesterase 2
|
|
chr9_-_115118145
Show fit
|
0.28 |
ENST00000350763.9
|
TNC
|
tenascin C
|
|
chr1_-_32901330
Show fit
|
0.28 |
ENST00000329151.5
ENST00000373463.8
|
TMEM54
|
transmembrane protein 54
|
|
chr17_-_4739866
Show fit
|
0.28 |
ENST00000574412.6
ENST00000293778.12
|
CXCL16
|
C-X-C motif chemokine ligand 16
|
|
chr9_-_115118198
Show fit
|
0.28 |
ENST00000534839.1
ENST00000535648.5
|
TNC
|
tenascin C
|
|
chr11_+_102317542
Show fit
|
0.28 |
ENST00000532808.5
|
BIRC3
|
baculoviral IAP repeat containing 3
|
|
chr11_+_72114840
Show fit
|
0.28 |
ENST00000622388.4
|
FOLR3
|
folate receptor gamma
|
|
chr6_+_32154010
Show fit
|
0.27 |
ENST00000375137.6
|
PPT2
|
palmitoyl-protein thioesterase 2
|
|
chr2_+_60881515
Show fit
|
0.26 |
ENST00000295025.12
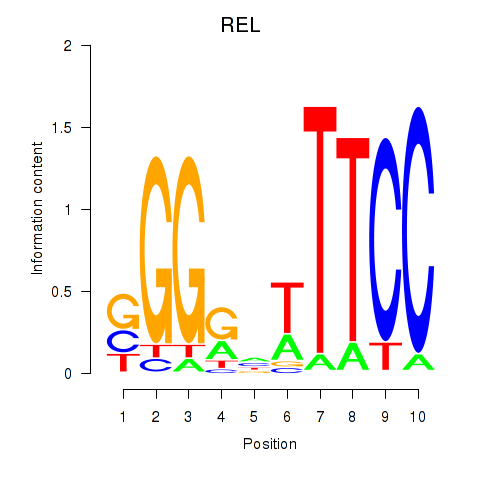
|
REL
|
REL proto-oncogene, NF-kB subunit
|
|
chr7_-_19145306
Show fit
|
0.26 |
ENST00000275461.3
|
FERD3L
|
Fer3 like bHLH transcription factor
|
|
chr10_-_107164692
Show fit
|
0.25 |
ENST00000263054.11
|
SORCS1
|
sortilin related VPS10 domain containing receptor 1
|
|
chr13_+_79481468
Show fit
|
0.25 |
ENST00000620924.1
|
NDFIP2
|
Nedd4 family interacting protein 2
|
|
chr11_+_102317450
Show fit
|
0.24 |
ENST00000615299.4
ENST00000527309.2
ENST00000526421.6
ENST00000263464.9
|
BIRC3
|
baculoviral IAP repeat containing 3
|
|
chr20_+_44355692
Show fit
|
0.24 |
ENST00000316673.8
ENST00000609795.5
ENST00000457232.5
ENST00000609262.5
|
HNF4A
|
hepatocyte nuclear factor 4 alpha
|
|
chr1_+_6206077
Show fit
|
0.24 |
ENST00000377939.5
|
RNF207
|
ring finger protein 207
|
|
chr10_-_94362925
Show fit
|
0.24 |
ENST00000371361.3
|
NOC3L
|
NOC3 like DNA replication regulator
|
|
chr13_+_79481446
Show fit
|
0.24 |
ENST00000487865.5
|
NDFIP2
|
Nedd4 family interacting protein 2
|
|
chr14_+_67619911
Show fit
|
0.24 |
ENST00000261783.4
|
ARG2
|
arginase 2
|
|
chr2_+_60881553
Show fit
|
0.24 |
ENST00000394479.4
|
REL
|
REL proto-oncogene, NF-kB subunit
|
|
chr6_-_29559724
Show fit
|
0.23 |
ENST00000377050.5
|
UBD
|
ubiquitin D
|
|
chr4_-_73998669
Show fit
|
0.22 |
ENST00000296027.5
|
CXCL5
|
C-X-C motif chemokine ligand 5
|
|
chr1_-_155990062
Show fit
|
0.22 |
ENST00000462460.6
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor 2
|
|
chr12_-_117099384
Show fit
|
0.22 |
ENST00000335209.12
|
TESC
|
tescalcin
|
|
chr12_-_117099434
Show fit
|
0.22 |
ENST00000541210.5
|
TESC
|
tescalcin
|
|
chr9_-_34637800
Show fit
|
0.21 |
ENST00000680730.1
ENST00000477726.1
|
SIGMAR1
|
sigma non-opioid intracellular receptor 1
|
|
chr19_+_40717091
Show fit
|
0.21 |
ENST00000263370.3
|
ITPKC
|
inositol-trisphosphate 3-kinase C
|
|
chr14_+_102777555
Show fit
|
0.21 |
ENST00000539721.5
ENST00000560463.5
|
TRAF3
|
TNF receptor associated factor 3
|
|
chr10_+_102394488
Show fit
|
0.21 |
ENST00000369966.8
|
NFKB2
|
nuclear factor kappa B subunit 2
|
|
chr2_-_88947820
Show fit
|
0.20 |
ENST00000496168.1
|
IGKV1-5
|
immunoglobulin kappa variable 1-5
|
|
chr14_-_68979436
Show fit
|
0.20 |
ENST00000193403.10
|
ACTN1
|
actinin alpha 1
|
|
chr14_+_103123452
Show fit
|
0.20 |
ENST00000558056.1
ENST00000560869.6
|
TNFAIP2
|
TNF alpha induced protein 2
|
|
chr6_+_150721073
Show fit
|
0.20 |
ENST00000358517.6
|
PLEKHG1
|
pleckstrin homology and RhoGEF domain containing G1
|
|
chr1_+_18630839
Show fit
|
0.20 |
ENST00000420770.7
|
PAX7
|
paired box 7
|
|
chr6_-_31582415
Show fit
|
0.19 |
ENST00000429299.3
ENST00000446745.2
|
LTB
|
lymphotoxin beta
|
|
chr16_-_75464655
Show fit
|
0.19 |
ENST00000569276.1
ENST00000357613.8
ENST00000561878.2
ENST00000566980.1
ENST00000567194.5
|
TMEM170A
ENSG00000261717.5
|
transmembrane protein 170A
novel TMEM170A-CFDP1 readthrough protein
|
|
chr12_+_11649666
Show fit
|
0.19 |
ENST00000396373.9
|
ETV6
|
ETS variant transcription factor 6
|
|
chr13_-_51453015
Show fit
|
0.19 |
ENST00000442263.4
ENST00000311234.9
|
INTS6
|
integrator complex subunit 6
|
|
chr4_-_74038681
Show fit
|
0.19 |
ENST00000296026.4
|
CXCL3
|
C-X-C motif chemokine ligand 3
|
|
chrX_-_40735476
Show fit
|
0.19 |
ENST00000324817.6
|
MED14
|
mediator complex subunit 14
|
|
chr2_+_90038848
Show fit
|
0.19 |
ENST00000390270.2
|
IGKV3D-20
|
immunoglobulin kappa variable 3D-20
|
|
chr21_-_41953997
Show fit
|
0.19 |
ENST00000380486.4
|
C2CD2
|
C2 calcium dependent domain containing 2
|
|
chr1_+_156147350
Show fit
|
0.18 |
ENST00000435124.5
ENST00000633494.1
|
SEMA4A
|
semaphorin 4A
|
|
chr7_-_99144053
Show fit
|
0.17 |
ENST00000361125.1
ENST00000361368.7
|
SMURF1
|
SMAD specific E3 ubiquitin protein ligase 1
|
|
chr2_-_219229309
Show fit
|
0.17 |
ENST00000443140.5
ENST00000432520.5
ENST00000409618.5
|
ATG9A
|
autophagy related 9A
|
|
chr2_-_74441882
Show fit
|
0.17 |
ENST00000272430.10
|
RTKN
|
rhotekin
|
|
chr15_+_41838839
Show fit
|
0.17 |
ENST00000458483.4
|
PLA2G4B
|
phospholipase A2 group IVB
|
|
chr3_-_53046031
Show fit
|
0.17 |
ENST00000482396.5
ENST00000394752.8
|
SFMBT1
|
Scm like with four mbt domains 1
|
|
chr16_-_75464364
Show fit
|
0.17 |
ENST00000569540.5
ENST00000566594.1
|
TMEM170A
ENSG00000261717.5
|
transmembrane protein 170A
novel TMEM170A-CFDP1 readthrough protein
|
|
chr2_-_219229571
Show fit
|
0.17 |
ENST00000436856.5
ENST00000428226.5
ENST00000409422.5
ENST00000361242.9
ENST00000431715.5
ENST00000457841.5
ENST00000439812.5
ENST00000396761.6
|
ATG9A
|
autophagy related 9A
|
|
chr2_-_162318613
Show fit
|
0.17 |
ENST00000649979.2
ENST00000421365.2
|
IFIH1
|
interferon induced with helicase C domain 1
|
|
chr12_-_122500947
Show fit
|
0.17 |
ENST00000672018.1
|
ZCCHC8
|
zinc finger CCHC-type containing 8
|
|
chr12_-_122500924
Show fit
|
0.17 |
ENST00000633063.3
|
ZCCHC8
|
zinc finger CCHC-type containing 8
|
|
chr1_-_155991198
Show fit
|
0.16 |
ENST00000673475.1
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor 2
|
|
chr12_+_57216779
Show fit
|
0.16 |
ENST00000349394.6
|
NXPH4
|
neurexophilin 4
|
|
chr12_+_55818033
Show fit
|
0.16 |
ENST00000552672.5
ENST00000243045.10
ENST00000550836.1
|
ORMDL2
|
ORMDL sphingolipid biosynthesis regulator 2
|
|
chr6_+_135181268
Show fit
|
0.16 |
ENST00000341911.10
ENST00000442647.7
ENST00000618728.4
ENST00000316528.12
ENST00000616088.4
|
MYB
|
MYB proto-oncogene, transcription factor
|
|
chr19_+_38899946
Show fit
|
0.16 |
ENST00000572515.5
ENST00000313582.6
ENST00000575359.5
|
NFKBIB
|
NFKB inhibitor beta
|
|
chr3_+_184249621
Show fit
|
0.16 |
ENST00000324557.9
ENST00000402825.7
|
EEF1AKMT4
EEF1AKMT4-ECE2
|
EEF1A lysine methyltransferase 4
EEF1AKMT4-ECE2 readthrough
|
|
chr19_-_45768843
Show fit
|
0.15 |
ENST00000560168.1
|
SIX5
|
SIX homeobox 5
|
|
chr11_-_105023136
Show fit
|
0.15 |
ENST00000526056.5
ENST00000531367.5
ENST00000456094.1
ENST00000444749.6
ENST00000393141.6
ENST00000418434.5
ENST00000260315.8
|
CASP5
|
caspase 5
|
|
chr5_-_150412743
Show fit
|
0.15 |
ENST00000353334.11
ENST00000009530.12
ENST00000377795.7
|
CD74
|
CD74 molecule
|
|
chr2_-_74465162
Show fit
|
0.15 |
ENST00000649854.1
ENST00000650523.1
ENST00000649601.1
ENST00000448666.7
ENST00000409065.5
ENST00000414701.1
ENST00000452063.7
ENST00000649075.1
ENST00000648810.1
ENST00000462443.2
|
MOGS
|
mannosyl-oligosaccharide glucosidase
|
|
chr18_-_46964408
Show fit
|
0.15 |
ENST00000676383.1
|
ENSG00000288616.1
|
elongin A3 family member D
|
|
chr3_-_49104745
Show fit
|
0.15 |
ENST00000635194.1
ENST00000306125.12
ENST00000634602.1
ENST00000414533.5
ENST00000635443.1
ENST00000452739.5
ENST00000635231.1
|
QARS1
|
glutaminyl-tRNA synthetase 1
|
|
chr2_+_15940537
Show fit
|
0.14 |
ENST00000281043.4
ENST00000638417.1
|
MYCN
|
MYCN proto-oncogene, bHLH transcription factor
|
|
chr22_-_33572227
Show fit
|
0.14 |
ENST00000674780.1
|
LARGE1
|
LARGE xylosyl- and glucuronyltransferase 1
|
|
chr3_-_184249520
Show fit
|
0.14 |
ENST00000455059.5
ENST00000445626.6
|
ALG3
|
ALG3 alpha-1,3- mannosyltransferase
|
|
chr19_+_8209320
Show fit
|
0.14 |
ENST00000561053.5
ENST00000559450.5
ENST00000251363.10
ENST00000559336.5
|
CERS4
|
ceramide synthase 4
|
|
chr2_+_90114838
Show fit
|
0.14 |
ENST00000417279.3
|
IGKV3D-15
|
immunoglobulin kappa variable 3D-15
|
|
chr12_+_6946468
Show fit
|
0.14 |
ENST00000543115.5
ENST00000399448.5
|
PTPN6
|
protein tyrosine phosphatase non-receptor type 6
|
|
chr20_-_45348414
Show fit
|
0.14 |
ENST00000372733.3
|
SDC4
|
syndecan 4
|
|
chr11_-_72752376
Show fit
|
0.14 |
ENST00000393609.8
|
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1
|
|
chr22_-_37984534
Show fit
|
0.13 |
ENST00000396884.8
|
SOX10
|
SRY-box transcription factor 10
|
|
chr14_+_77181780
Show fit
|
0.13 |
ENST00000298351.5
ENST00000554346.5
|
TMEM63C
|
transmembrane protein 63C
|
|
chr19_-_48746797
Show fit
|
0.13 |
ENST00000602105.1
ENST00000332955.7
|
IZUMO1
|
izumo sperm-egg fusion 1
|
|
chr5_+_177086867
Show fit
|
0.13 |
ENST00000503708.5
ENST00000393648.6
ENST00000514472.1
ENST00000502906.5
ENST00000292408.9
ENST00000510911.5
|
FGFR4
|
fibroblast growth factor receptor 4
|
|
chr15_-_73368951
Show fit
|
0.13 |
ENST00000261917.4
|
HCN4
|
hyperpolarization activated cyclic nucleotide gated potassium channel 4
|
|
chr14_+_102777461
Show fit
|
0.13 |
ENST00000560371.5
ENST00000347662.8
|
TRAF3
|
TNF receptor associated factor 3
|
|
chr11_-_66546021
Show fit
|
0.13 |
ENST00000310442.5
|
ZDHHC24
|
zinc finger DHHC-type containing 24
|
|
chr7_+_156949704
Show fit
|
0.12 |
ENST00000275820.4
|
NOM1
|
nucleolar protein with MIF4G domain 1
|
|
chr19_+_4229502
Show fit
|
0.12 |
ENST00000221847.6
|
EBI3
|
Epstein-Barr virus induced 3
|
|
chr19_+_38899680
Show fit
|
0.12 |
ENST00000576510.5
ENST00000392079.7
|
NFKBIB
|
NFKB inhibitor beta
|
|
chr3_-_52897541
Show fit
|
0.12 |
ENST00000355083.11
ENST00000504329.1
|
STIMATE
STIMATE-MUSTN1
|
STIM activating enhancer
STIMATE-MUSTN1 readthrough
|
|
chr9_-_133479075
Show fit
|
0.12 |
ENST00000414172.1
ENST00000371897.8
ENST00000371899.9
|
SLC2A6
|
solute carrier family 2 member 6
|
|
chr4_-_76036060
Show fit
|
0.12 |
ENST00000306621.8
|
CXCL11
|
C-X-C motif chemokine ligand 11
|
|
chr19_+_33373694
Show fit
|
0.12 |
ENST00000284000.9
|
CEBPG
|
CCAAT enhancer binding protein gamma
|
|
chr1_+_63367619
Show fit
|
0.12 |
ENST00000263440.6
|
ALG6
|
ALG6 alpha-1,3-glucosyltransferase
|
|
chr14_-_68979314
Show fit
|
0.12 |
ENST00000684713.1
ENST00000683198.1
ENST00000684598.1
ENST00000682331.1
ENST00000682291.1
ENST00000683342.1
|
ACTN1
|
actinin alpha 1
|
|
chr4_+_73836667
Show fit
|
0.12 |
ENST00000226317.10
|
CXCL6
|
C-X-C motif chemokine ligand 6
|
|
chr3_-_49104457
Show fit
|
0.12 |
ENST00000635292.1
ENST00000635541.1
ENST00000464962.6
|
QARS1
|
glutaminyl-tRNA synthetase 1
|
|
chr3_+_47803125
Show fit
|
0.11 |
ENST00000445061.6
ENST00000348968.8
|
DHX30
|
DExH-box helicase 30
|
|
chr16_+_84175933
Show fit
|
0.11 |
ENST00000569735.1
|
DNAAF1
|
dynein axonemal assembly factor 1
|
|
chr10_-_73808616
Show fit
|
0.11 |
ENST00000299641.8
|
NDST2
|
N-deacetylase and N-sulfotransferase 2
|
|
chrX_+_153642473
Show fit
|
0.11 |
ENST00000370167.8
|
DUSP9
|
dual specificity phosphatase 9
|
|
chr17_+_51260520
Show fit
|
0.11 |
ENST00000225298.12
|
UTP18
|
UTP18 small subunit processome component
|
|
chr1_+_202010575
Show fit
|
0.11 |
ENST00000367283.7
ENST00000367284.10
|
ELF3
|
E74 like ETS transcription factor 3
|
|
chr14_+_102777433
Show fit
|
0.11 |
ENST00000392745.8
|
TRAF3
|
TNF receptor associated factor 3
|
|
chr3_+_101561833
Show fit
|
0.11 |
ENST00000309922.7
|
TRMT10C
|
tRNA methyltransferase 10C, mitochondrial RNase P subunit
|
|
chr4_-_55546900
Show fit
|
0.11 |
ENST00000513440.6
|
CLOCK
|
clock circadian regulator
|
|
chr2_-_113235443
Show fit
|
0.11 |
ENST00000465084.1
|
PAX8
|
paired box 8
|
|
chr22_-_27919199
Show fit
|
0.11 |
ENST00000634017.1
ENST00000436663.1
ENST00000320996.14
ENST00000335272.10
ENST00000455418.7
|
PITPNB
|
phosphatidylinositol transfer protein beta
|
|
chr6_+_10694916
Show fit
|
0.11 |
ENST00000379568.4
|
PAK1IP1
|
PAK1 interacting protein 1
|
|
chr6_-_142946312
Show fit
|
0.11 |
ENST00000367604.6
|
HIVEP2
|
HIVEP zinc finger 2
|
|
chr9_-_136304084
Show fit
|
0.10 |
ENST00000638797.2
ENST00000624277.3
|
CCDC187
|
coiled-coil domain containing 187
|
|
chr16_-_2196575
Show fit
|
0.10 |
ENST00000343516.8
|
CASKIN1
|
CASK interacting protein 1
|
|
chr2_+_218880844
Show fit
|
0.10 |
ENST00000258411.8
|
WNT10A
|
Wnt family member 10A
|
|
chr7_+_69599588
Show fit
|
0.10 |
ENST00000403018.3
|
AUTS2
|
activator of transcription and developmental regulator AUTS2
|
|
chr3_+_169911566
Show fit
|
0.10 |
ENST00000428432.6
ENST00000335556.7
|
SAMD7
|
sterile alpha motif domain containing 7
|
|
chr2_-_68467272
Show fit
|
0.10 |
ENST00000377957.4
|
FBXO48
|
F-box protein 48
|
|
chr16_+_67537439
Show fit
|
0.09 |
ENST00000428437.6
ENST00000569253.5
|
RIPOR1
|
RHO family interacting cell polarization regulator 1
|
|
chr20_-_37527862
Show fit
|
0.09 |
ENST00000373537.7
ENST00000445723.5
ENST00000414080.1
|
BLCAP
|
BLCAP apoptosis inducing factor
|
|
chr3_+_101561926
Show fit
|
0.09 |
ENST00000495642.1
|
TRMT10C
|
tRNA methyltransferase 10C, mitochondrial RNase P subunit
|
|
chr10_+_104641282
Show fit
|
0.09 |
ENST00000369701.8
|
SORCS3
|
sortilin related VPS10 domain containing receptor 3
|
|
chr16_+_68843515
Show fit
|
0.09 |
ENST00000261778.2
|
TANGO6
|
transport and golgi organization 6 homolog
|
|
chr1_-_61725121
Show fit
|
0.09 |
ENST00000371177.2
ENST00000606498.5
|
TM2D1
|
TM2 domain containing 1
|
|
chr5_+_50667405
Show fit
|
0.09 |
ENST00000505554.5
|
PARP8
|
poly(ADP-ribose) polymerase family member 8
|
|
chr14_+_37197904
Show fit
|
0.09 |
ENST00000556615.5
ENST00000396294.6
|
MIPOL1
|
mirror-image polydactyly 1
|
|
chr14_+_37197921
Show fit
|
0.09 |
ENST00000684589.1
ENST00000327441.11
|
MIPOL1
|
mirror-image polydactyly 1
|
|
chr2_+_108377947
Show fit
|
0.09 |
ENST00000272452.7
|
SULT1C4
|
sulfotransferase family 1C member 4
|
|
chr4_-_184474518
Show fit
|
0.08 |
ENST00000393593.8
|
IRF2
|
interferon regulatory factor 2
|
|
chr14_+_37197988
Show fit
|
0.08 |
ENST00000539062.6
|
MIPOL1
|
mirror-image polydactyly 1
|
|
chr11_+_119168188
Show fit
|
0.08 |
ENST00000454811.5
ENST00000409265.8
ENST00000449394.5
|
NLRX1
|
NLR family member X1
|
|
chr12_-_39619782
Show fit
|
0.08 |
ENST00000308666.4
|
ABCD2
|
ATP binding cassette subfamily D member 2
|
|
chr2_+_201182873
Show fit
|
0.08 |
ENST00000360132.7
|
CASP10
|
caspase 10
|
|
chr4_+_113292925
Show fit
|
0.08 |
ENST00000673353.1
ENST00000505342.6
ENST00000672915.1
ENST00000509550.5
|
ANK2
|
ankyrin 2
|
|
chr20_-_37527723
Show fit
|
0.08 |
ENST00000397135.1
ENST00000397137.5
|
BLCAP
|
BLCAP apoptosis inducing factor
|
|
chr2_-_28870266
Show fit
|
0.08 |
ENST00000306108.10
|
TRMT61B
|
tRNA methyltransferase 61B
|
|
chr3_+_101827982
Show fit
|
0.08 |
ENST00000461724.5
ENST00000483180.5
ENST00000394054.6
|
NFKBIZ
|
NFKB inhibitor zeta
|
|
chr15_+_85380625
Show fit
|
0.08 |
ENST00000560302.5
|
AKAP13
|
A-kinase anchoring protein 13
|
|
chr5_+_96936071
Show fit
|
0.08 |
ENST00000231368.10
|
LNPEP
|
leucyl and cystinyl aminopeptidase
|
|
chrX_+_116436599
Show fit
|
0.08 |
ENST00000598581.3
|
SLC6A14
|
solute carrier family 6 member 14
|
|
chr9_+_6328363
Show fit
|
0.07 |
ENST00000344545.6
ENST00000381428.1
ENST00000314556.4
|
TPD52L3
|
TPD52 like 3
|
|
chr4_+_113292838
Show fit
|
0.07 |
ENST00000672411.1
ENST00000673231.1
|
ANK2
|
ankyrin 2
|
|
chr2_+_201183120
Show fit
|
0.07 |
ENST00000272879.9
ENST00000286186.11
ENST00000374650.7
ENST00000346817.9
ENST00000313728.11
ENST00000448480.1
|
CASP10
|
caspase 10
|
|
chr4_+_72031902
Show fit
|
0.07 |
ENST00000344413.6
ENST00000308744.12
|
NPFFR2
|
neuropeptide FF receptor 2
|
|
chr16_+_50696999
Show fit
|
0.07 |
ENST00000300589.6
|
NOD2
|
nucleotide binding oligomerization domain containing 2
|
|
chr13_-_98577131
Show fit
|
0.07 |
ENST00000397517.6
|
STK24
|
serine/threonine kinase 24
|
|
chr22_+_46335699
Show fit
|
0.07 |
ENST00000645190.1
ENST00000642562.1
ENST00000381019.3
|
TRMU
|
tRNA mitochondrial 2-thiouridylase
|
|
chr21_+_33266350
Show fit
|
0.07 |
ENST00000290200.7
|
IL10RB
|
interleukin 10 receptor subunit beta
|
|
chr10_+_30434176
Show fit
|
0.07 |
ENST00000263056.6
ENST00000375322.2
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8
|
|
chr14_+_37198025
Show fit
|
0.07 |
ENST00000556451.5
ENST00000556753.5
|
MIPOL1
|
mirror-image polydactyly 1
|
|
chr19_+_3721719
Show fit
|
0.07 |
ENST00000589378.5
|
TJP3
|
tight junction protein 3
|
|
chr16_+_84176067
Show fit
|
0.07 |
ENST00000564928.1
|
DNAAF1
|
dynein axonemal assembly factor 1
|
|
chr15_+_76059973
Show fit
|
0.07 |
ENST00000388942.8
|
TMEM266
|
transmembrane protein 266
|
|
chr20_-_37527891
Show fit
|
0.07 |
ENST00000414542.6
|
BLCAP
|
BLCAP apoptosis inducing factor
|
|
chr4_-_145180496
Show fit
|
0.07 |
ENST00000447906.8
|
OTUD4
|
OTU deubiquitinase 4
|
|
chr16_+_24256313
Show fit
|
0.06 |
ENST00000005284.4
|
CACNG3
|
calcium voltage-gated channel auxiliary subunit gamma 3
|
|
chr11_-_65614195
Show fit
|
0.06 |
ENST00000309100.8
ENST00000529839.1
ENST00000526293.1
|
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11
|
|
chr3_+_9890567
Show fit
|
0.06 |
ENST00000489724.2
|
JAGN1
|
jagunal homolog 1
|
|
chr1_+_202416826
Show fit
|
0.06 |
ENST00000466968.1
|
PPP1R12B
|
protein phosphatase 1 regulatory subunit 12B
|
|
chr2_-_18560616
Show fit
|
0.06 |
ENST00000381249.4
|
RDH14
|
retinol dehydrogenase 14
|
|
chr10_+_35605337
Show fit
|
0.06 |
ENST00000321660.2
|
GJD4
|
gap junction protein delta 4
|
|
chr11_-_128522189
Show fit
|
0.06 |
ENST00000526145.6
|
ETS1
|
ETS proto-oncogene 1, transcription factor
|