Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
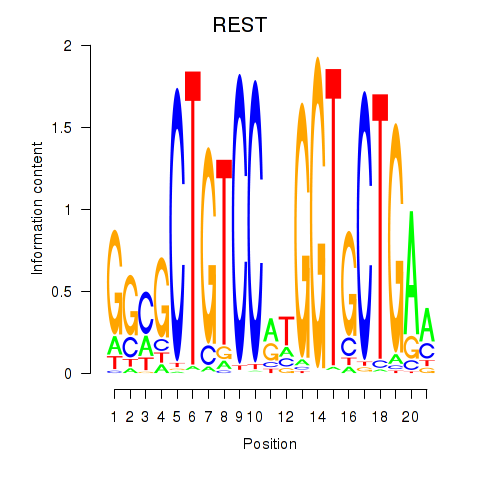
Results for REST
Z-value: 0.02
Transcription factors associated with REST
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
REST
|
ENSG00000084093.19 | RE1 silencing transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| REST | hg38_v1_chr4_+_56908894_56909016, hg38_v1_chr4_+_56907876_56907947 | -0.56 | 1.5e-01 | Click! |
Activity profile of REST motif
Sorted Z-values of REST motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_155070317 | 0.22 |
ENST00000287907.3
|
HTR5A
|
5-hydroxytryptamine receptor 5A |
| chr1_+_209938207 | 0.14 |
ENST00000472886.5
|
SYT14
|
synaptotagmin 14 |
| chr12_+_71439976 | 0.13 |
ENST00000536515.5
ENST00000540815.2 |
LGR5
|
leucine rich repeat containing G protein-coupled receptor 5 |
| chr1_+_209938169 | 0.13 |
ENST00000367019.5
ENST00000537238.5 ENST00000637265.1 |
SYT14
|
synaptotagmin 14 |
| chr12_+_71439789 | 0.11 |
ENST00000266674.10
|
LGR5
|
leucine rich repeat containing G protein-coupled receptor 5 |
| chr9_-_101594995 | 0.11 |
ENST00000636434.1
|
PPP3R2
|
protein phosphatase 3 regulatory subunit B, beta |
| chr17_-_3433841 | 0.10 |
ENST00000248384.1
|
OR1E2
|
olfactory receptor family 1 subfamily E member 2 |
| chr2_-_47570905 | 0.09 |
ENST00000327876.5
|
KCNK12
|
potassium two pore domain channel subfamily K member 12 |
| chr20_+_5911501 | 0.08 |
ENST00000378961.9
ENST00000455042.1 |
CHGB
|
chromogranin B |
| chr11_+_20599602 | 0.08 |
ENST00000525748.6
|
SLC6A5
|
solute carrier family 6 member 5 |
| chr12_+_113358542 | 0.08 |
ENST00000545182.6
ENST00000280800.5 |
PLBD2
|
phospholipase B domain containing 2 |
| chr14_+_92923143 | 0.08 |
ENST00000216492.10
ENST00000334654.4 |
CHGA
|
chromogranin A |
| chr15_-_72320149 | 0.07 |
ENST00000287202.10
|
CELF6
|
CUGBP Elav-like family member 6 |
| chr17_+_32486975 | 0.07 |
ENST00000313401.4
|
CDK5R1
|
cyclin dependent kinase 5 regulatory subunit 1 |
| chr3_+_194136138 | 0.07 |
ENST00000232424.4
|
HES1
|
hes family bHLH transcription factor 1 |
| chr1_+_154567820 | 0.06 |
ENST00000637900.1
|
CHRNB2
|
cholinergic receptor nicotinic beta 2 subunit |
| chr11_-_6655788 | 0.06 |
ENST00000299441.5
|
DCHS1
|
dachsous cadherin-related 1 |
| chr3_+_120908072 | 0.06 |
ENST00000273666.10
ENST00000471454.6 ENST00000472879.5 ENST00000497029.5 ENST00000492541.5 |
STXBP5L
|
syntaxin binding protein 5 like |
| chr11_-_125903190 | 0.06 |
ENST00000227474.8
ENST00000613398.4 ENST00000534158.5 ENST00000529801.1 |
PUS3
|
pseudouridine synthase 3 |
| chr11_+_62613236 | 0.05 |
ENST00000278833.4
|
ROM1
|
retinal outer segment membrane protein 1 |
| chr11_+_92969651 | 0.05 |
ENST00000257068.3
ENST00000528076.1 |
MTNR1B
|
melatonin receptor 1B |
| chr16_+_285680 | 0.05 |
ENST00000435833.1
|
PDIA2
|
protein disulfide isomerase family A member 2 |
| chr11_-_93197932 | 0.05 |
ENST00000326402.9
|
SLC36A4
|
solute carrier family 36 member 4 |
| chrX_-_49200174 | 0.05 |
ENST00000472598.5
ENST00000263233.9 ENST00000479808.5 |
SYP
|
synaptophysin |
| chr14_+_93430927 | 0.05 |
ENST00000393151.6
|
UNC79
|
unc-79 homolog, NALCN channel complex subunit |
| chr9_+_132582581 | 0.05 |
ENST00000263610.7
|
BARHL1
|
BarH like homeobox 1 |
| chr2_-_51032390 | 0.05 |
ENST00000404971.5
|
NRXN1
|
neurexin 1 |
| chrX_+_7147675 | 0.05 |
ENST00000674429.1
|
STS
|
steroid sulfatase |
| chr19_+_39268394 | 0.05 |
ENST00000331982.6
|
IFNL2
|
interferon lambda 2 |
| chr11_+_637244 | 0.05 |
ENST00000176183.6
|
DRD4
|
dopamine receptor D4 |
| chr2_+_225399684 | 0.05 |
ENST00000636099.1
|
NYAP2
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2 |
| chr8_-_90082871 | 0.05 |
ENST00000265431.7
|
CALB1
|
calbindin 1 |
| chrX_+_7147237 | 0.04 |
ENST00000666110.2
|
STS
|
steroid sulfatase |
| chr9_-_101594918 | 0.04 |
ENST00000374806.2
|
PPP3R2
|
protein phosphatase 3 regulatory subunit B, beta |
| chr2_-_51032151 | 0.04 |
ENST00000628515.2
|
NRXN1
|
neurexin 1 |
| chr3_-_114179052 | 0.04 |
ENST00000383673.5
ENST00000295881.9 |
DRD3
|
dopamine receptor D3 |
| chr16_+_2969270 | 0.04 |
ENST00000293978.12
|
PAQR4
|
progestin and adipoQ receptor family member 4 |
| chr10_-_101031095 | 0.04 |
ENST00000645349.1
ENST00000619208.6 ENST00000644782.1 ENST00000370215.7 ENST00000646029.1 |
PDZD7
|
PDZ domain containing 7 |
| chr16_+_2969307 | 0.03 |
ENST00000576565.1
ENST00000318782.9 |
PAQR4
|
progestin and adipoQ receptor family member 4 |
| chr5_+_175871570 | 0.03 |
ENST00000512824.5
ENST00000393745.8 |
CPLX2
|
complexin 2 |
| chr12_-_29783798 | 0.03 |
ENST00000552618.5
ENST00000551659.5 ENST00000539277.6 |
TMTC1
|
transmembrane O-mannosyltransferase targeting cadherins 1 |
| chr9_-_133609325 | 0.03 |
ENST00000673969.1
|
FAM163B
|
family with sequence similarity 163 member B |
| chr2_-_51032091 | 0.03 |
ENST00000637511.1
ENST00000405472.7 ENST00000405581.3 ENST00000401669.7 |
NRXN1
|
neurexin 1 |
| chr12_-_57050102 | 0.03 |
ENST00000300119.8
|
MYO1A
|
myosin IA |
| chr2_-_27308445 | 0.03 |
ENST00000296099.2
|
UCN
|
urocortin |
| chr6_+_154039102 | 0.03 |
ENST00000360422.8
ENST00000330432.12 |
OPRM1
|
opioid receptor mu 1 |
| chr3_-_114178698 | 0.03 |
ENST00000467632.5
|
DRD3
|
dopamine receptor D3 |
| chr1_+_32886456 | 0.03 |
ENST00000373467.4
|
HPCA
|
hippocalcin |
| chr1_+_37793865 | 0.03 |
ENST00000397631.7
|
MANEAL
|
mannosidase endo-alpha like |
| chr14_+_93430853 | 0.03 |
ENST00000553484.5
|
UNC79
|
unc-79 homolog, NALCN channel complex subunit |
| chr1_-_151716782 | 0.03 |
ENST00000420342.1
ENST00000290583.9 |
CELF3
|
CUGBP Elav-like family member 3 |
| chr17_+_58148384 | 0.03 |
ENST00000268912.6
ENST00000641449.1 |
OR4D1
|
olfactory receptor family 4 subfamily D member 1 |
| chr8_+_6708626 | 0.03 |
ENST00000285518.11
|
AGPAT5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 |
| chr4_-_174829212 | 0.03 |
ENST00000340217.5
ENST00000274093.8 |
GLRA3
|
glycine receptor alpha 3 |
| chr19_+_40530464 | 0.03 |
ENST00000392023.1
|
SPTBN4
|
spectrin beta, non-erythrocytic 4 |
| chr2_+_130756291 | 0.03 |
ENST00000458606.5
ENST00000423981.1 |
AMER3
|
APC membrane recruitment protein 3 |
| chr12_+_66755 | 0.03 |
ENST00000538872.6
|
IQSEC3
|
IQ motif and Sec7 domain ArfGEF 3 |
| chr19_-_55180010 | 0.02 |
ENST00000589172.5
|
SYT5
|
synaptotagmin 5 |
| chr6_+_158536398 | 0.02 |
ENST00000367090.4
|
TMEM181
|
transmembrane protein 181 |
| chr3_+_75906666 | 0.02 |
ENST00000487694.7
ENST00000602589.5 |
ROBO2
|
roundabout guidance receptor 2 |
| chr10_-_49762335 | 0.02 |
ENST00000419399.4
ENST00000432695.2 |
OGDHL
|
oxoglutarate dehydrogenase L |
| chr1_-_101996919 | 0.02 |
ENST00000370103.9
|
OLFM3
|
olfactomedin 3 |
| chr3_+_62319037 | 0.02 |
ENST00000494481.5
|
C3orf14
|
chromosome 3 open reading frame 14 |
| chr19_-_3971051 | 0.02 |
ENST00000545797.7
ENST00000596311.5 |
DAPK3
|
death associated protein kinase 3 |
| chr1_+_37793787 | 0.02 |
ENST00000373045.11
|
MANEAL
|
mannosidase endo-alpha like |
| chr1_+_212432891 | 0.02 |
ENST00000366988.5
|
NENF
|
neudesin neurotrophic factor |
| chr17_-_3398410 | 0.02 |
ENST00000322608.2
|
OR1E1
|
olfactory receptor family 1 subfamily E member 1 |
| chr3_+_62318983 | 0.02 |
ENST00000232519.9
ENST00000462069.6 ENST00000465142.5 |
C3orf14
|
chromosome 3 open reading frame 14 |
| chr4_+_157220654 | 0.02 |
ENST00000393815.6
|
GRIA2
|
glutamate ionotropic receptor AMPA type subunit 2 |
| chr10_-_44385043 | 0.02 |
ENST00000374426.6
ENST00000395794.2 ENST00000374429.6 ENST00000395793.7 ENST00000343575.10 ENST00000395795.5 |
CXCL12
|
C-X-C motif chemokine ligand 12 |
| chr10_-_49762276 | 0.02 |
ENST00000374103.9
|
OGDHL
|
oxoglutarate dehydrogenase L |
| chr9_-_121050264 | 0.02 |
ENST00000223642.3
|
C5
|
complement C5 |
| chr20_+_37383648 | 0.02 |
ENST00000373567.6
|
SRC
|
SRC proto-oncogene, non-receptor tyrosine kinase |
| chr9_+_135714582 | 0.01 |
ENST00000630792.2
|
KCNT1
|
potassium sodium-activated channel subfamily T member 1 |
| chr16_+_4958289 | 0.01 |
ENST00000251170.12
|
SEC14L5
|
SEC14 like lipid binding 5 |
| chr9_+_135714425 | 0.01 |
ENST00000473941.5
ENST00000486577.6 |
KCNT1
|
potassium sodium-activated channel subfamily T member 1 |
| chr5_+_71719267 | 0.01 |
ENST00000296777.5
|
CARTPT
|
CART prepropeptide |
| chr17_-_7315084 | 0.01 |
ENST00000570780.5
|
GPS2
|
G protein pathway suppressor 2 |
| chr2_-_51032536 | 0.01 |
ENST00000626899.1
ENST00000406316.6 |
NRXN1
|
neurexin 1 |
| chr10_+_59177161 | 0.01 |
ENST00000373878.3
|
PHYHIPL
|
phytanoyl-CoA 2-hydroxylase interacting protein like |
| chr4_+_157220691 | 0.01 |
ENST00000509417.5
ENST00000645636.1 ENST00000296526.12 ENST00000264426.14 |
GRIA2
|
glutamate ionotropic receptor AMPA type subunit 2 |
| chr11_+_45885625 | 0.01 |
ENST00000241014.6
|
MAPK8IP1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr16_+_31459950 | 0.01 |
ENST00000564900.1
|
ARMC5
|
armadillo repeat containing 5 |
| chr6_+_109978256 | 0.01 |
ENST00000414000.3
|
GPR6
|
G protein-coupled receptor 6 |
| chr1_+_154567770 | 0.01 |
ENST00000368476.4
|
CHRNB2
|
cholinergic receptor nicotinic beta 2 subunit |
| chr1_-_175743529 | 0.01 |
ENST00000367674.7
ENST00000263525.6 |
TNR
|
tenascin R |
| chrX_-_47619850 | 0.01 |
ENST00000295987.13
ENST00000340666.5 |
SYN1
|
synapsin I |
| chr16_+_24256313 | 0.01 |
ENST00000005284.4
|
CACNG3
|
calcium voltage-gated channel auxiliary subunit gamma 3 |
| chr18_-_43277482 | 0.01 |
ENST00000255224.8
ENST00000590752.5 ENST00000596867.1 |
SYT4
|
synaptotagmin 4 |
| chr21_-_32728030 | 0.01 |
ENST00000382499.7
ENST00000433931.7 ENST00000382491.7 |
SYNJ1
|
synaptojanin 1 |
| chr6_+_109978297 | 0.01 |
ENST00000275169.5
|
GPR6
|
G protein-coupled receptor 6 |
| chr7_+_8433602 | 0.00 |
ENST00000405863.6
|
NXPH1
|
neurexophilin 1 |
| chr14_-_94293071 | 0.00 |
ENST00000554723.5
|
SERPINA10
|
serpin family A member 10 |
| chr14_-_94293024 | 0.00 |
ENST00000393096.5
|
SERPINA10
|
serpin family A member 10 |
| chr14_-_94293260 | 0.00 |
ENST00000261994.9
|
SERPINA10
|
serpin family A member 10 |
| chr3_-_132035004 | 0.00 |
ENST00000429747.6
|
CPNE4
|
copine 4 |
| chr6_+_138404206 | 0.00 |
ENST00000607197.6
ENST00000367697.7 |
HEBP2
|
heme binding protein 2 |
| chr3_-_48662877 | 0.00 |
ENST00000164024.5
|
CELSR3
|
cadherin EGF LAG seven-pass G-type receptor 3 |
| chrX_-_153971810 | 0.00 |
ENST00000310441.12
|
HCFC1
|
host cell factor C1 |
| chr1_+_171781635 | 0.00 |
ENST00000361735.4
ENST00000362019.7 ENST00000367737.9 |
EEF1AKNMT
|
eEF1A lysine and N-terminal methyltransferase |
| chr9_+_137139481 | 0.00 |
ENST00000371546.8
ENST00000371550.8 ENST00000371553.7 ENST00000371555.8 ENST00000371559.8 ENST00000371560.4 |
GRIN1
|
glutamate ionotropic receptor NMDA type subunit 1 |
| chr5_-_151924824 | 0.00 |
ENST00000455880.2
|
GLRA1
|
glycine receptor alpha 1 |
| chr17_+_44308573 | 0.00 |
ENST00000590941.5
ENST00000225441.11 ENST00000426726.8 |
RUNDC3A
|
RUN domain containing 3A |
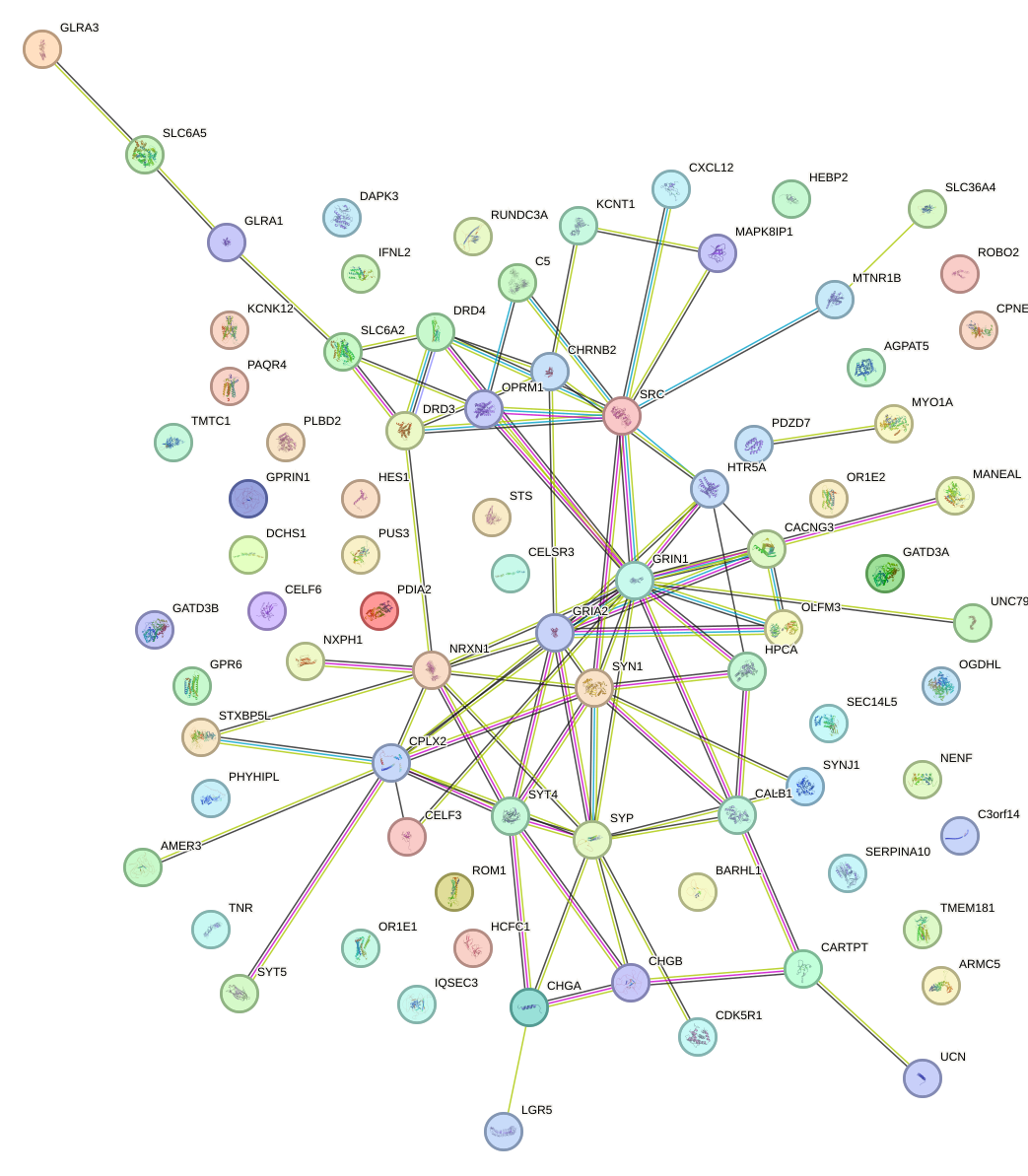
Network of associatons between targets according to the STRING database.
First level regulatory network of REST
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.1 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.1 | GO:1901899 | positive regulation of relaxation of cardiac muscle(GO:1901899) |
| 0.0 | 0.1 | GO:0021722 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.0 | 0.1 | GO:0060164 | trochlear nerve development(GO:0021558) auditory receptor cell fate determination(GO:0042668) negative regulation of auditory receptor cell differentiation(GO:0045608) regulation of timing of neuron differentiation(GO:0060164) negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.0 | 0.1 | GO:0050883 | musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 | 0.2 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.0 | GO:0051944 | positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.0 | 0.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.0 | GO:0035483 | gastric motility(GO:0035482) gastric emptying(GO:0035483) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.0 | 0.1 | GO:0004773 | steryl-sulfatase activity(GO:0004773) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.2 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |