Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
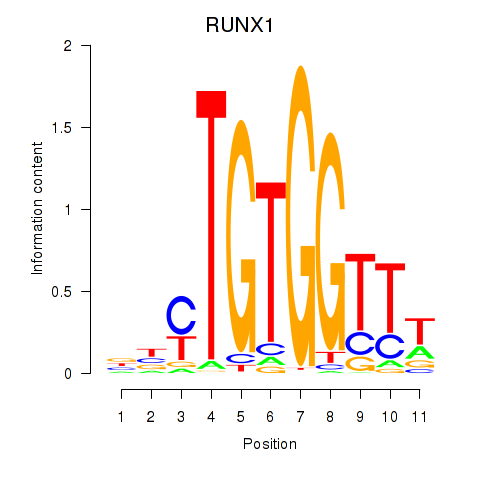
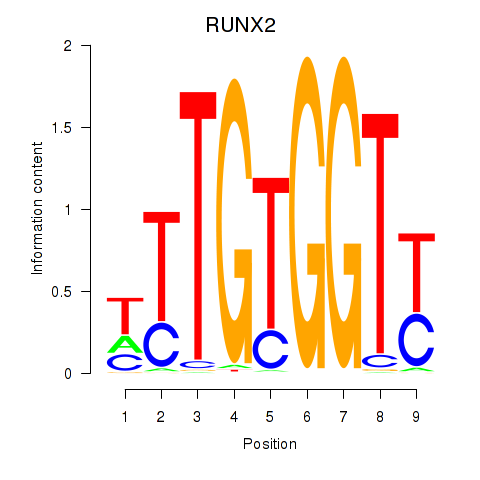
Results for RUNX1_RUNX2
Z-value: 0.31
Transcription factors associated with RUNX1_RUNX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RUNX1
|
ENSG00000159216.19 | RUNX family transcription factor 1 |
|
RUNX2
|
ENSG00000124813.23 | RUNX family transcription factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RUNX1 | hg38_v1_chr21_-_34888683_34888714 | 0.28 | 5.0e-01 | Click! |
| RUNX2 | hg38_v1_chr6_+_45422485_45422561 | -0.22 | 6.0e-01 | Click! |
Activity profile of RUNX1_RUNX2 motif
Sorted Z-values of RUNX1_RUNX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_48978313 | 0.31 |
ENST00000293549.4
|
WNT1
|
Wnt family member 1 |
| chr19_-_6720641 | 0.28 |
ENST00000245907.11
|
C3
|
complement C3 |
| chr1_+_86424154 | 0.27 |
ENST00000370565.5
|
CLCA2
|
chloride channel accessory 2 |
| chr2_+_168802563 | 0.26 |
ENST00000445023.6
|
NOSTRIN
|
nitric oxide synthase trafficking |
| chr2_+_168802610 | 0.26 |
ENST00000397206.6
ENST00000317647.12 ENST00000397209.6 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr3_-_190122317 | 0.24 |
ENST00000427335.6
|
P3H2
|
prolyl 3-hydroxylase 2 |
| chr12_+_57583101 | 0.20 |
ENST00000674858.1
ENST00000675433.1 ENST00000674980.1 |
KIF5A
|
kinesin family member 5A |
| chr2_+_37344594 | 0.20 |
ENST00000404976.5
ENST00000338415.8 |
QPCT
|
glutaminyl-peptide cyclotransferase |
| chr5_+_132073782 | 0.20 |
ENST00000296871.4
|
CSF2
|
colony stimulating factor 2 |
| chr17_-_36196748 | 0.19 |
ENST00000619989.1
|
CCL3L1
|
C-C motif chemokine ligand 3 like 1 |
| chr8_-_19682576 | 0.19 |
ENST00000332246.10
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr18_+_13611910 | 0.18 |
ENST00000590308.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr9_-_127778659 | 0.17 |
ENST00000314830.13
|
SH2D3C
|
SH2 domain containing 3C |
| chr4_+_70226116 | 0.17 |
ENST00000317987.6
|
FDCSP
|
follicular dendritic cell secreted protein |
| chr6_+_26500296 | 0.17 |
ENST00000684113.1
|
BTN1A1
|
butyrophilin subfamily 1 member A1 |
| chr12_-_55707865 | 0.17 |
ENST00000347027.10
ENST00000257879.11 ENST00000553804.6 |
ITGA7
|
integrin subunit alpha 7 |
| chr5_+_76403266 | 0.16 |
ENST00000274364.11
|
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr17_-_36090133 | 0.16 |
ENST00000613922.2
|
CCL3
|
C-C motif chemokine ligand 3 |
| chr20_+_46008900 | 0.16 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 |
| chr11_-_1750558 | 0.15 |
ENST00000340134.5
|
IFITM10
|
interferon induced transmembrane protein 10 |
| chr11_+_121576760 | 0.15 |
ENST00000532694.5
ENST00000534286.5 |
SORL1
|
sortilin related receptor 1 |
| chr18_-_55422306 | 0.15 |
ENST00000566777.5
ENST00000626584.2 |
TCF4
|
transcription factor 4 |
| chr14_+_22497657 | 0.15 |
ENST00000390496.1
|
TRAJ41
|
T cell receptor alpha joining 41 |
| chr19_-_54242751 | 0.14 |
ENST00000245621.6
ENST00000396365.6 |
LILRA6
|
leukocyte immunoglobulin like receptor A6 |
| chr15_+_73683938 | 0.14 |
ENST00000567189.5
|
CD276
|
CD276 molecule |
| chr1_+_152514474 | 0.14 |
ENST00000368790.4
|
CRCT1
|
cysteine rich C-terminal 1 |
| chr2_-_218985176 | 0.14 |
ENST00000295727.2
|
FEV
|
FEV transcription factor, ETS family member |
| chr4_-_83114715 | 0.14 |
ENST00000426923.2
ENST00000311507.9 ENST00000509973.5 |
PLAC8
|
placenta associated 8 |
| chr7_+_144000320 | 0.14 |
ENST00000641698.1
|
OR6B1
|
olfactory receptor family 6 subfamily B member 1 |
| chr16_+_30375820 | 0.13 |
ENST00000566955.1
|
MYLPF
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr18_+_23949847 | 0.13 |
ENST00000588004.1
|
LAMA3
|
laminin subunit alpha 3 |
| chr2_+_127418420 | 0.13 |
ENST00000234071.8
ENST00000429925.5 ENST00000442644.5 |
PROC
|
protein C, inactivator of coagulation factors Va and VIIIa |
| chr16_-_66918839 | 0.13 |
ENST00000565235.2
ENST00000568632.5 ENST00000565796.5 |
CDH16
|
cadherin 16 |
| chr6_-_33080710 | 0.13 |
ENST00000419277.5
|
HLA-DPA1
|
major histocompatibility complex, class II, DP alpha 1 |
| chr16_-_66918876 | 0.13 |
ENST00000570262.5
ENST00000299752.9 ENST00000394055.7 |
CDH16
|
cadherin 16 |
| chr1_+_200894892 | 0.13 |
ENST00000413687.3
|
INAVA
|
innate immunity activator |
| chr1_+_236394268 | 0.13 |
ENST00000334232.9
|
EDARADD
|
EDAR associated death domain |
| chr17_-_58417547 | 0.13 |
ENST00000577716.5
|
RNF43
|
ring finger protein 43 |
| chr17_-_58417521 | 0.13 |
ENST00000584437.5
ENST00000407977.7 |
RNF43
|
ring finger protein 43 |
| chr1_-_23369813 | 0.12 |
ENST00000314011.9
|
ZNF436
|
zinc finger protein 436 |
| chr10_-_62816341 | 0.12 |
ENST00000242480.4
ENST00000637191.1 |
EGR2
|
early growth response 2 |
| chr16_+_85611401 | 0.12 |
ENST00000405402.6
|
GSE1
|
Gse1 coiled-coil protein |
| chr6_+_72216745 | 0.12 |
ENST00000517827.5
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr6_-_29045175 | 0.12 |
ENST00000377175.2
|
OR2W1
|
olfactory receptor family 2 subfamily W member 1 |
| chr17_-_41586887 | 0.12 |
ENST00000167586.7
|
KRT14
|
keratin 14 |
| chr3_+_45026296 | 0.12 |
ENST00000296130.5
|
CLEC3B
|
C-type lectin domain family 3 member B |
| chr7_-_29195186 | 0.12 |
ENST00000449801.5
ENST00000409850.5 |
CPVL
|
carboxypeptidase vitellogenic like |
| chr6_-_29375291 | 0.12 |
ENST00000396806.3
|
OR12D3
|
olfactory receptor family 12 subfamily D member 3 |
| chr10_-_13234329 | 0.12 |
ENST00000463405.2
|
UCMA
|
upper zone of growth plate and cartilage matrix associated |
| chr18_-_55422492 | 0.12 |
ENST00000561992.5
ENST00000630712.2 |
TCF4
|
transcription factor 4 |
| chr10_-_13234368 | 0.12 |
ENST00000378681.8
|
UCMA
|
upper zone of growth plate and cartilage matrix associated |
| chr10_+_80413817 | 0.12 |
ENST00000372187.9
|
PRXL2A
|
peroxiredoxin like 2A |
| chr6_-_106975309 | 0.11 |
ENST00000615659.1
|
CD24
|
CD24 molecule |
| chr20_-_1588632 | 0.11 |
ENST00000262929.9
ENST00000567028.5 |
SIRPB1
ENSG00000260861.6
|
signal regulatory protein beta 1 novel protein, SIRPB1-SIRPD readthrough |
| chr17_-_7261092 | 0.11 |
ENST00000574070.5
|
CLDN7
|
claudin 7 |
| chr18_+_31447732 | 0.11 |
ENST00000257189.5
|
DSG3
|
desmoglein 3 |
| chr11_+_1838970 | 0.11 |
ENST00000381911.6
|
TNNI2
|
troponin I2, fast skeletal type |
| chr15_-_43776956 | 0.11 |
ENST00000319359.8
|
ELL3
|
elongation factor for RNA polymerase II 3 |
| chr11_+_60971668 | 0.11 |
ENST00000313421.11
|
CD6
|
CD6 molecule |
| chr1_-_184037695 | 0.11 |
ENST00000361927.9
ENST00000649786.1 |
COLGALT2
|
collagen beta(1-O)galactosyltransferase 2 |
| chr14_-_24634266 | 0.11 |
ENST00000382540.5
|
GZMB
|
granzyme B |
| chr17_-_31314066 | 0.11 |
ENST00000577894.1
|
EVI2B
|
ecotropic viral integration site 2B |
| chr1_+_209686173 | 0.11 |
ENST00000615289.4
ENST00000367028.6 ENST00000261465.5 |
HSD11B1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr19_-_51372640 | 0.11 |
ENST00000600427.5
ENST00000221978.10 |
NKG7
|
natural killer cell granule protein 7 |
| chr14_-_24634160 | 0.10 |
ENST00000616551.1
ENST00000382542.5 ENST00000216341.9 ENST00000526004.1 ENST00000415355.7 |
GZMB
|
granzyme B |
| chr6_+_31575557 | 0.10 |
ENST00000449264.3
|
TNF
|
tumor necrosis factor |
| chr15_-_93089192 | 0.10 |
ENST00000329082.11
|
RGMA
|
repulsive guidance molecule BMP co-receptor a |
| chr18_+_68798065 | 0.10 |
ENST00000360242.9
|
CCDC102B
|
coiled-coil domain containing 102B |
| chr19_-_43465596 | 0.10 |
ENST00000244333.4
|
LYPD3
|
LY6/PLAUR domain containing 3 |
| chr10_-_104085847 | 0.10 |
ENST00000648076.2
|
COL17A1
|
collagen type XVII alpha 1 chain |
| chr19_+_17470474 | 0.10 |
ENST00000598424.5
ENST00000252595.12 |
SLC27A1
|
solute carrier family 27 member 1 |
| chr11_+_113908983 | 0.10 |
ENST00000537778.5
|
HTR3B
|
5-hydroxytryptamine receptor 3B |
| chr11_-_102530738 | 0.10 |
ENST00000260227.5
|
MMP7
|
matrix metallopeptidase 7 |
| chr22_+_46620380 | 0.10 |
ENST00000406902.6
|
GRAMD4
|
GRAM domain containing 4 |
| chr6_+_72216442 | 0.10 |
ENST00000425662.6
ENST00000453976.6 |
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr2_+_232662733 | 0.10 |
ENST00000410095.5
ENST00000611312.1 |
EFHD1
|
EF-hand domain family member D1 |
| chr11_-_118264445 | 0.10 |
ENST00000438295.2
|
MPZL2
|
myelin protein zero like 2 |
| chr3_-_151316795 | 0.10 |
ENST00000260843.5
|
GPR87
|
G protein-coupled receptor 87 |
| chr1_-_56966006 | 0.10 |
ENST00000371237.9
|
C8B
|
complement C8 beta chain |
| chr10_+_128047559 | 0.10 |
ENST00000306042.9
|
PTPRE
|
protein tyrosine phosphatase receptor type E |
| chr15_+_83447411 | 0.10 |
ENST00000324537.5
|
SH3GL3
|
SH3 domain containing GRB2 like 3, endophilin A3 |
| chr11_+_1868673 | 0.10 |
ENST00000405957.6
|
LSP1
|
lymphocyte specific protein 1 |
| chr7_+_75398915 | 0.10 |
ENST00000437796.1
|
TRIM73
|
tripartite motif containing 73 |
| chr19_+_41262656 | 0.10 |
ENST00000599719.5
ENST00000601309.5 |
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U like 1 |
| chr5_-_150086511 | 0.10 |
ENST00000675795.1
|
CSF1R
|
colony stimulating factor 1 receptor |
| chr2_+_112977998 | 0.10 |
ENST00000259205.5
ENST00000376489.6 |
IL36G
|
interleukin 36 gamma |
| chrX_-_48971829 | 0.10 |
ENST00000218176.4
|
KCND1
|
potassium voltage-gated channel subfamily D member 1 |
| chr3_-_49903863 | 0.09 |
ENST00000296474.8
ENST00000621387.4 |
MST1R
|
macrophage stimulating 1 receptor |
| chr2_-_216694794 | 0.09 |
ENST00000449583.1
|
IGFBP5
|
insulin like growth factor binding protein 5 |
| chr10_+_110226805 | 0.09 |
ENST00000651495.1
ENST00000652506.1 ENST00000651811.1 ENST00000651167.1 ENST00000651516.1 ENST00000651467.1 ENST00000651004.1 ENST00000650843.1 ENST00000650644.1 ENST00000650696.1 ENST00000652604.1 ENST00000652463.1 ENST00000650810.1 ENST00000650952.1 ENST00000652028.1 ENST00000651866.1 ENST00000651848.1 ENST00000442296.5 ENST00000369612.1 |
MXI1
|
MAX interactor 1, dimerization protein |
| chr22_+_24607638 | 0.09 |
ENST00000432867.5
|
GGT1
|
gamma-glutamyltransferase 1 |
| chr2_-_162318129 | 0.09 |
ENST00000679938.1
|
IFIH1
|
interferon induced with helicase C domain 1 |
| chr11_-_111923722 | 0.09 |
ENST00000527950.5
|
CRYAB
|
crystallin alpha B |
| chr17_-_7294592 | 0.09 |
ENST00000007699.10
|
YBX2
|
Y-box binding protein 2 |
| chr1_-_56966133 | 0.09 |
ENST00000535057.5
ENST00000543257.5 |
C8B
|
complement C8 beta chain |
| chr19_-_49155130 | 0.09 |
ENST00000595625.1
|
HRC
|
histidine rich calcium binding protein |
| chr4_+_75556048 | 0.09 |
ENST00000616557.1
ENST00000435974.2 ENST00000311623.9 |
ODAPH
|
odontogenesis associated phosphoprotein |
| chr20_-_1619996 | 0.09 |
ENST00000381603.7
ENST00000381605.9 ENST00000279477.11 ENST00000568365.1 ENST00000564763.1 |
SIRPB1
ENSG00000260861.6
|
signal regulatory protein beta 1 novel protein, SIRPB1-SIRPD readthrough |
| chr12_-_85836372 | 0.09 |
ENST00000361228.5
|
RASSF9
|
Ras association domain family member 9 |
| chr11_+_2902258 | 0.09 |
ENST00000649076.2
ENST00000449793.6 |
SLC22A18
|
solute carrier family 22 member 18 |
| chr2_-_221572272 | 0.09 |
ENST00000409854.5
ENST00000443796.5 ENST00000281821.7 |
EPHA4
|
EPH receptor A4 |
| chr15_+_73684204 | 0.09 |
ENST00000537340.6
ENST00000318424.9 |
CD276
|
CD276 molecule |
| chr5_+_145936554 | 0.09 |
ENST00000359120.9
|
SH3RF2
|
SH3 domain containing ring finger 2 |
| chr11_+_1853049 | 0.09 |
ENST00000311604.8
|
LSP1
|
lymphocyte specific protein 1 |
| chr19_-_43780957 | 0.09 |
ENST00000648319.1
|
KCNN4
|
potassium calcium-activated channel subfamily N member 4 |
| chr17_-_31314040 | 0.09 |
ENST00000330927.5
|
EVI2B
|
ecotropic viral integration site 2B |
| chr4_-_158159657 | 0.09 |
ENST00000590648.5
|
GASK1B
|
golgi associated kinase 1B |
| chr4_+_87975829 | 0.09 |
ENST00000614857.5
|
SPP1
|
secreted phosphoprotein 1 |
| chr5_+_170861990 | 0.09 |
ENST00000523189.6
|
RANBP17
|
RAN binding protein 17 |
| chr17_-_42050587 | 0.09 |
ENST00000587304.3
|
C17orf113
|
chromosome 17 open reading frame 113 |
| chr12_+_25052512 | 0.09 |
ENST00000557489.5
ENST00000354454.7 ENST00000536173.5 |
IRAG2
|
inositol 1,4,5-triphosphate receptor associated 2 |
| chr1_-_3611470 | 0.08 |
ENST00000356575.9
|
MEGF6
|
multiple EGF like domains 6 |
| chr7_-_141946926 | 0.08 |
ENST00000438351.1
ENST00000439991.1 ENST00000551012.6 ENST00000546910.6 |
CLEC5A
|
C-type lectin domain containing 5A |
| chr17_+_19378476 | 0.08 |
ENST00000395604.8
ENST00000482850.1 |
MAPK7
|
mitogen-activated protein kinase 7 |
| chr7_+_100969565 | 0.08 |
ENST00000536621.6
ENST00000379442.7 |
MUC12
|
mucin 12, cell surface associated |
| chr16_+_56932134 | 0.08 |
ENST00000439977.7
ENST00000300302.9 ENST00000344114.8 ENST00000379792.6 |
HERPUD1
|
homocysteine inducible ER protein with ubiquitin like domain 1 |
| chr3_-_151203201 | 0.08 |
ENST00000480322.1
ENST00000309180.6 |
GPR171
|
G protein-coupled receptor 171 |
| chr11_+_76783349 | 0.08 |
ENST00000333090.5
|
TSKU
|
tsukushi, small leucine rich proteoglycan |
| chr2_+_74458400 | 0.08 |
ENST00000393972.7
ENST00000233615.7 ENST00000409737.5 ENST00000428943.1 |
WBP1
|
WW domain binding protein 1 |
| chr2_-_89143133 | 0.08 |
ENST00000492167.1
|
IGKV3-20
|
immunoglobulin kappa variable 3-20 |
| chr12_+_29223730 | 0.08 |
ENST00000547116.5
|
FAR2
|
fatty acyl-CoA reductase 2 |
| chr6_-_106974721 | 0.08 |
ENST00000606017.2
ENST00000620405.1 |
CD24
|
CD24 molecule |
| chr4_+_87975667 | 0.08 |
ENST00000237623.11
ENST00000682655.1 ENST00000508233.6 ENST00000360804.4 ENST00000395080.8 |
SPP1
|
secreted phosphoprotein 1 |
| chr1_+_196774813 | 0.08 |
ENST00000471440.6
ENST00000391985.7 ENST00000617219.1 ENST00000367425.9 |
CFHR3
|
complement factor H related 3 |
| chr12_+_29223659 | 0.08 |
ENST00000182377.8
|
FAR2
|
fatty acyl-CoA reductase 2 |
| chr4_-_185775271 | 0.08 |
ENST00000430503.5
ENST00000319454.10 ENST00000450341.5 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr6_+_26440472 | 0.08 |
ENST00000494393.5
ENST00000482451.5 ENST00000471353.5 ENST00000361232.7 ENST00000487627.5 ENST00000496719.1 ENST00000244519.7 ENST00000490254.5 ENST00000487272.1 |
BTN3A3
|
butyrophilin subfamily 3 member A3 |
| chr3_+_98763331 | 0.08 |
ENST00000485391.5
ENST00000492254.5 |
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr13_-_28100556 | 0.08 |
ENST00000241453.12
|
FLT3
|
fms related receptor tyrosine kinase 3 |
| chr1_-_237890922 | 0.08 |
ENST00000611898.4
ENST00000366570.5 |
ZP4
|
zona pellucida glycoprotein 4 |
| chr1_+_111229813 | 0.08 |
ENST00000524472.5
|
CHI3L2
|
chitinase 3 like 2 |
| chr2_+_219460719 | 0.08 |
ENST00000396688.5
|
SPEG
|
striated muscle enriched protein kinase |
| chr16_-_87936529 | 0.08 |
ENST00000649794.3
ENST00000649158.1 ENST00000648177.1 |
CA5A
|
carbonic anhydrase 5A |
| chr10_+_97584314 | 0.08 |
ENST00000370647.8
|
HOGA1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr10_+_89392546 | 0.08 |
ENST00000546318.2
ENST00000371804.4 |
IFIT1
|
interferon induced protein with tetratricopeptide repeats 1 |
| chr11_-_120138104 | 0.08 |
ENST00000341846.10
|
TRIM29
|
tripartite motif containing 29 |
| chr11_-_16356538 | 0.08 |
ENST00000683767.1
|
SOX6
|
SRY-box transcription factor 6 |
| chr19_-_8502621 | 0.08 |
ENST00000600262.1
ENST00000423345.5 |
PRAM1
|
PML-RARA regulated adaptor molecule 1 |
| chr6_-_137219028 | 0.07 |
ENST00000647124.1
ENST00000642390.1 ENST00000645753.1 ENST00000646036.1 ENST00000646898.1 ENST00000644894.1 |
IFNGR1
|
interferon gamma receptor 1 |
| chr15_-_78937198 | 0.07 |
ENST00000677207.1
|
CTSH
|
cathepsin H |
| chr2_+_101998955 | 0.07 |
ENST00000393414.6
|
IL1R2
|
interleukin 1 receptor type 2 |
| chr21_+_41361999 | 0.07 |
ENST00000436410.5
ENST00000435611.6 ENST00000330714.8 |
MX2
|
MX dynamin like GTPase 2 |
| chrX_-_66639022 | 0.07 |
ENST00000374719.8
|
EDA2R
|
ectodysplasin A2 receptor |
| chr7_+_29194757 | 0.07 |
ENST00000222792.11
|
CHN2
|
chimerin 2 |
| chr17_+_7455571 | 0.07 |
ENST00000575379.1
|
CHRNB1
|
cholinergic receptor nicotinic beta 1 subunit |
| chr4_-_142474395 | 0.07 |
ENST00000507861.5
|
INPP4B
|
inositol polyphosphate-4-phosphatase type II B |
| chr12_+_25052634 | 0.07 |
ENST00000548766.5
|
IRAG2
|
inositol 1,4,5-triphosphate receptor associated 2 |
| chr1_-_223363337 | 0.07 |
ENST00000608996.5
|
SUSD4
|
sushi domain containing 4 |
| chr9_-_92404559 | 0.07 |
ENST00000262551.8
ENST00000375561.10 |
OGN
|
osteoglycin |
| chr1_-_24964984 | 0.07 |
ENST00000338888.3
ENST00000399916.5 |
RUNX3
|
RUNX family transcription factor 3 |
| chr11_+_124611420 | 0.07 |
ENST00000284288.3
|
PANX3
|
pannexin 3 |
| chr5_-_150113344 | 0.07 |
ENST00000286301.7
ENST00000511344.1 |
CSF1R
|
colony stimulating factor 1 receptor |
| chrX_+_48683763 | 0.07 |
ENST00000376701.5
|
WAS
|
WASP actin nucleation promoting factor |
| chr4_+_88378842 | 0.07 |
ENST00000264346.12
|
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr12_-_70637405 | 0.07 |
ENST00000548122.2
ENST00000551525.5 ENST00000550358.5 ENST00000334414.11 |
PTPRB
|
protein tyrosine phosphatase receptor type B |
| chr21_-_46605073 | 0.07 |
ENST00000291700.9
ENST00000367071.4 |
S100B
|
S100 calcium binding protein B |
| chr7_+_117020191 | 0.07 |
ENST00000434836.5
ENST00000393443.5 ENST00000465133.5 ENST00000477742.5 ENST00000393444.7 ENST00000393447.8 |
ST7
|
suppression of tumorigenicity 7 |
| chr15_+_43594027 | 0.07 |
ENST00000453733.5
ENST00000441322.6 ENST00000627381.1 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr3_-_99850976 | 0.07 |
ENST00000487087.5
|
FILIP1L
|
filamin A interacting protein 1 like |
| chr20_+_63861498 | 0.07 |
ENST00000369916.5
|
ABHD16B
|
abhydrolase domain containing 16B |
| chr11_+_108223027 | 0.07 |
ENST00000675843.1
ENST00000683468.1 ENST00000532931.5 ENST00000530958.5 |
ATM
|
ATM serine/threonine kinase |
| chr14_-_24508251 | 0.07 |
ENST00000250378.7
ENST00000206446.4 |
CMA1
|
chymase 1 |
| chr11_+_64340191 | 0.07 |
ENST00000356786.10
|
CCDC88B
|
coiled-coil domain containing 88B |
| chr11_+_7605719 | 0.07 |
ENST00000530181.5
|
PPFIBP2
|
PPFIA binding protein 2 |
| chr4_+_88378733 | 0.07 |
ENST00000273960.7
ENST00000380265.9 |
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr11_+_36375978 | 0.07 |
ENST00000378867.7
|
PRR5L
|
proline rich 5 like |
| chr15_+_83447328 | 0.07 |
ENST00000427482.7
|
SH3GL3
|
SH3 domain containing GRB2 like 3, endophilin A3 |
| chr8_+_133113483 | 0.07 |
ENST00000521107.1
|
TG
|
thyroglobulin |
| chr17_-_41521719 | 0.07 |
ENST00000393976.6
|
KRT15
|
keratin 15 |
| chr18_+_36544544 | 0.06 |
ENST00000591635.5
|
FHOD3
|
formin homology 2 domain containing 3 |
| chr4_+_105895458 | 0.06 |
ENST00000379987.7
|
NPNT
|
nephronectin |
| chr20_+_44714853 | 0.06 |
ENST00000372865.4
|
CCN5
|
cellular communication network factor 5 |
| chr9_+_34653864 | 0.06 |
ENST00000556792.5
ENST00000318041.13 |
IL11RA
|
interleukin 11 receptor subunit alpha |
| chr16_-_10694922 | 0.06 |
ENST00000283025.7
|
TEKT5
|
tektin 5 |
| chr1_+_6448448 | 0.06 |
ENST00000475228.6
|
ESPN
|
espin |
| chr16_-_58294976 | 0.06 |
ENST00000543437.5
ENST00000569079.1 |
PRSS54
|
serine protease 54 |
| chrX_-_78659328 | 0.06 |
ENST00000321110.2
|
RTL3
|
retrotransposon Gag like 3 |
| chr6_+_89081787 | 0.06 |
ENST00000354922.3
|
PNRC1
|
proline rich nuclear receptor coactivator 1 |
| chr16_-_58295019 | 0.06 |
ENST00000567164.6
ENST00000219301.8 ENST00000569727.1 |
PRSS54
|
serine protease 54 |
| chr4_+_165873711 | 0.06 |
ENST00000513213.5
|
TLL1
|
tolloid like 1 |
| chr1_+_111229692 | 0.06 |
ENST00000466741.5
ENST00000477185.6 |
CHI3L2
|
chitinase 3 like 2 |
| chr16_+_82035245 | 0.06 |
ENST00000199936.9
|
HSD17B2
|
hydroxysteroid 17-beta dehydrogenase 2 |
| chrX_-_151974668 | 0.06 |
ENST00000370328.4
|
GABRE
|
gamma-aminobutyric acid type A receptor subunit epsilon |
| chr20_+_44714835 | 0.06 |
ENST00000372868.6
|
CCN5
|
cellular communication network factor 5 |
| chr6_+_88047822 | 0.06 |
ENST00000237201.2
|
SPACA1
|
sperm acrosome associated 1 |
| chr10_+_102461380 | 0.06 |
ENST00000238936.8
ENST00000369931.3 |
MFSD13A
|
major facilitator superfamily domain containing 13A |
| chr5_-_43412323 | 0.06 |
ENST00000361115.4
|
CCL28
|
C-C motif chemokine ligand 28 |
| chr17_-_74546082 | 0.06 |
ENST00000330793.2
|
CD300C
|
CD300c molecule |
| chrX_-_47574738 | 0.06 |
ENST00000640721.1
|
SYN1
|
synapsin I |
| chr5_+_35856883 | 0.06 |
ENST00000506850.5
ENST00000303115.8 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr14_+_104745960 | 0.06 |
ENST00000556623.1
ENST00000555674.1 |
ADSS1
|
adenylosuccinate synthase 1 |
| chr2_-_230960954 | 0.06 |
ENST00000392039.2
|
GPR55
|
G protein-coupled receptor 55 |
| chr1_+_153774210 | 0.06 |
ENST00000271857.6
|
SLC27A3
|
solute carrier family 27 member 3 |
| chr4_+_105895435 | 0.06 |
ENST00000453617.6
ENST00000427316.6 ENST00000514622.5 ENST00000305572.12 |
NPNT
|
nephronectin |
| chr18_+_63907948 | 0.06 |
ENST00000238508.8
|
SERPINB10
|
serpin family B member 10 |
| chr17_+_19377721 | 0.06 |
ENST00000308406.9
ENST00000299612.11 |
MAPK7
|
mitogen-activated protein kinase 7 |
| chr6_-_48111132 | 0.06 |
ENST00000398738.3
ENST00000679966.1 ENST00000339488.9 |
PTCHD4
|
patched domain containing 4 |
| chr19_+_39997031 | 0.06 |
ENST00000599504.5
ENST00000596894.5 ENST00000601138.5 ENST00000347077.9 ENST00000600094.5 |
ZNF546
|
zinc finger protein 546 |
| chr14_-_94388589 | 0.06 |
ENST00000402629.1
ENST00000556091.1 ENST00000393087.9 ENST00000554720.1 |
SERPINA1
|
serpin family A member 1 |
| chr19_-_49155384 | 0.06 |
ENST00000252825.9
|
HRC
|
histidine rich calcium binding protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of RUNX1_RUNX2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0061184 | positive regulation of dermatome development(GO:0061184) |
| 0.1 | 0.3 | GO:0001796 | type IIa hypersensitivity(GO:0001794) regulation of type IIa hypersensitivity(GO:0001796) type II hypersensitivity(GO:0002445) regulation of type II hypersensitivity(GO:0002892) |
| 0.1 | 0.2 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.1 | 0.2 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.1 | 0.3 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.2 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.1 | GO:1902948 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.0 | 0.1 | GO:1901899 | positive regulation of relaxation of cardiac muscle(GO:1901899) |
| 0.0 | 0.2 | GO:0032599 | B cell receptor transport within lipid bilayer(GO:0032595) B cell receptor transport into membrane raft(GO:0032597) protein transport out of membrane raft(GO:0032599) chemokine receptor transport out of membrane raft(GO:0032600) negative regulation of transforming growth factor beta3 production(GO:0032913) chemokine receptor transport within lipid bilayer(GO:0033606) |
| 0.0 | 0.1 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.0 | 0.2 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.0 | 0.1 | GO:0021593 | rhombomere morphogenesis(GO:0021593) rhombomere 3 morphogenesis(GO:0021658) |
| 0.0 | 0.1 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.0 | 0.1 | GO:0030806 | negative regulation of cyclic nucleotide catabolic process(GO:0030806) negative regulation of cAMP catabolic process(GO:0030821) |
| 0.0 | 0.1 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.0 | 0.1 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.0 | 0.1 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.1 | GO:1904808 | regulation of protein oxidation(GO:1904806) positive regulation of protein oxidation(GO:1904808) |
| 0.0 | 0.2 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.1 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.0 | 0.1 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.2 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.0 | 0.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.2 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.0 | 0.1 | GO:2000721 | pilomotor reflex(GO:0097195) positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.1 | GO:1904328 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.0 | 0.1 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 0.0 | 0.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.0 | 0.1 | GO:1904882 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.0 | 0.1 | GO:0050720 | interleukin-1 beta biosynthetic process(GO:0050720) |
| 0.0 | 0.1 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.1 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.2 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.0 | 0.1 | GO:1903904 | negative regulation of establishment of T cell polarity(GO:1903904) negative regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001107) |
| 0.0 | 0.1 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.1 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.1 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.0 | 0.1 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.0 | 0.1 | GO:1904761 | negative regulation of myofibroblast differentiation(GO:1904761) negative regulation of vascular smooth muscle cell differentiation(GO:1905064) |
| 0.0 | 0.3 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.1 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.1 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.0 | 0.0 | GO:2000538 | regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.1 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.0 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.0 | 0.1 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.1 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.0 | 0.1 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:0021779 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.0 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.0 | 0.0 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.0 | 0.1 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.0 | 0.0 | GO:0051695 | actin filament uncapping(GO:0051695) |
| 0.0 | 0.1 | GO:0051459 | regulation of corticotropin secretion(GO:0051459) |
| 0.0 | 0.1 | GO:0071104 | response to interleukin-9(GO:0071104) |
| 0.0 | 0.0 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 0.0 | 0.0 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.0 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.0 | 0.1 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.0 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.0 | 0.0 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.0 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.0 | 0.0 | GO:0070889 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) platelet alpha granule organization(GO:0070889) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.1 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.0 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.0 | 0.0 | GO:2001170 | negative regulation of mitochondrial DNA replication(GO:0090298) negative regulation of mitochondrial DNA metabolic process(GO:1901859) negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.0 | 0.0 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 0.0 | 0.0 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.0 | 0.0 | GO:0043000 | negative regulation of anion channel activity(GO:0010360) Golgi to plasma membrane CFTR protein transport(GO:0043000) |
| 0.0 | 0.0 | GO:1903216 | negative regulation of late endosome to lysosome transport(GO:1902823) regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 0.0 | 0.2 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.0 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.0 | 0.2 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0071754 | IgM immunoglobulin complex(GO:0071753) IgM immunoglobulin complex, circulating(GO:0071754) pentameric IgM immunoglobulin complex(GO:0071756) |
| 0.0 | 0.2 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.1 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.0 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.0 | 0.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.0 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.1 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.0 | 0.0 | GO:0044753 | amphisome(GO:0044753) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 0.1 | 0.2 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.1 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.0 | 0.2 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.2 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.0 | 0.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.4 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.2 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.0 | 0.1 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.1 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.1 | GO:0030395 | lactose binding(GO:0030395) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 0.1 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.3 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.0 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.0 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0017161 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.0 | 0.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.0 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.0 | 0.0 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.1 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.0 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.0 | 0.2 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.5 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.3 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |