Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for SMAD3
Z-value: 0.26
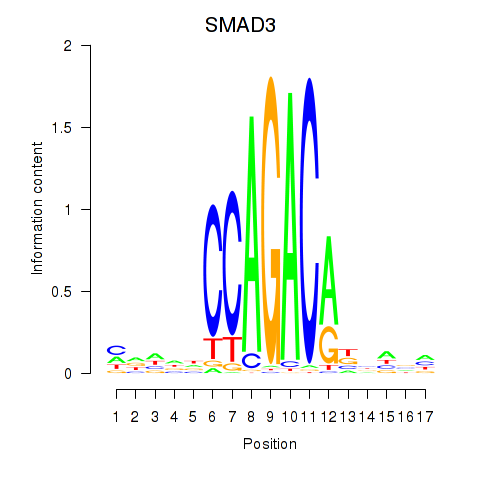
Transcription factors associated with SMAD3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SMAD3
|
ENSG00000166949.17 | SMAD family member 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SMAD3 | hg38_v1_chr15_+_67065586_67065614, hg38_v1_chr15_+_67067780_67067811 | 0.66 | 7.6e-02 | Click! |
Activity profile of SMAD3 motif
Sorted Z-values of SMAD3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_89838289 | 0.16 |
ENST00000336904.7
|
SNCA
|
synuclein alpha |
| chr20_+_59676661 | 0.13 |
ENST00000355648.8
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr19_-_55180010 | 0.12 |
ENST00000589172.5
|
SYT5
|
synaptotagmin 5 |
| chr12_+_3491189 | 0.12 |
ENST00000382622.4
|
PRMT8
|
protein arginine methyltransferase 8 |
| chr6_+_26124161 | 0.11 |
ENST00000377791.4
ENST00000602637.1 |
H2AC6
|
H2A clustered histone 6 |
| chr6_-_27132750 | 0.10 |
ENST00000607124.1
ENST00000339812.3 |
H2BC11
|
H2B clustered histone 11 |
| chr4_-_185775271 | 0.10 |
ENST00000430503.5
ENST00000319454.10 ENST00000450341.5 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr6_-_27831557 | 0.10 |
ENST00000611927.2
|
H4C12
|
H4 clustered histone 12 |
| chr6_-_27146841 | 0.09 |
ENST00000356950.2
|
H2BC12
|
H2B clustered histone 12 |
| chr3_+_12796662 | 0.09 |
ENST00000456430.6
ENST00000626378.1 |
CAND2
|
cullin associated and neddylation dissociated 2 (putative) |
| chr1_+_153416517 | 0.08 |
ENST00000368729.9
|
S100A7A
|
S100 calcium binding protein A7A |
| chr6_-_27807916 | 0.08 |
ENST00000377401.3
|
H2BC13
|
H2B clustered histone 13 |
| chr15_-_82647336 | 0.08 |
ENST00000617522.4
ENST00000684509.1 |
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr6_+_30882914 | 0.07 |
ENST00000509639.5
ENST00000412274.6 ENST00000507901.5 ENST00000507046.5 ENST00000437124.6 ENST00000454612.6 ENST00000396342.6 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr15_-_82709886 | 0.07 |
ENST00000666055.1
ENST00000261722.8 ENST00000535513.2 |
AP3B2
|
adaptor related protein complex 3 subunit beta 2 |
| chr9_-_133131109 | 0.07 |
ENST00000372062.8
|
RALGDS
|
ral guanine nucleotide dissociation stimulator |
| chr10_+_92691897 | 0.07 |
ENST00000492654.3
|
HHEX
|
hematopoietically expressed homeobox |
| chr6_-_27814757 | 0.07 |
ENST00000333151.5
|
H2AC14
|
H2A clustered histone 14 |
| chr15_+_81299416 | 0.06 |
ENST00000558332.3
|
IL16
|
interleukin 16 |
| chr6_+_27824084 | 0.06 |
ENST00000355057.3
|
H4C11
|
H4 clustered histone 11 |
| chr15_-_82709859 | 0.06 |
ENST00000542200.2
ENST00000535359.6 ENST00000668990.2 ENST00000652847.1 |
AP3B2
|
adaptor related protein complex 3 subunit beta 2 |
| chr17_+_50095331 | 0.06 |
ENST00000503176.6
|
PDK2
|
pyruvate dehydrogenase kinase 2 |
| chr15_-_82647503 | 0.06 |
ENST00000567678.1
ENST00000620182.4 |
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr6_+_27815010 | 0.06 |
ENST00000621112.2
|
H2BC14
|
H2B clustered histone 14 |
| chr7_+_134565098 | 0.06 |
ENST00000652743.1
|
AKR1B15
|
aldo-keto reductase family 1 member B15 |
| chr10_-_88952763 | 0.06 |
ENST00000224784.10
|
ACTA2
|
actin alpha 2, smooth muscle |
| chr6_-_11807045 | 0.05 |
ENST00000379415.6
|
ADTRP
|
androgen dependent TFPI regulating protein |
| chr3_+_125969172 | 0.05 |
ENST00000514116.6
ENST00000513830.5 |
ROPN1B
|
rhophilin associated tail protein 1B |
| chr6_-_26216673 | 0.05 |
ENST00000541790.3
|
H2BC8
|
H2B clustered histone 8 |
| chr6_+_25726767 | 0.05 |
ENST00000274764.5
|
H2BC1
|
H2B clustered histone 1 |
| chr15_-_82647960 | 0.05 |
ENST00000615198.4
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr6_+_122717544 | 0.05 |
ENST00000354275.2
ENST00000368446.1 |
PKIB
|
cAMP-dependent protein kinase inhibitor beta |
| chr16_-_58295019 | 0.05 |
ENST00000567164.6
ENST00000219301.8 ENST00000569727.1 |
PRSS54
|
serine protease 54 |
| chr19_-_11577632 | 0.05 |
ENST00000590420.1
ENST00000648477.1 |
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr16_-_58294976 | 0.05 |
ENST00000543437.5
ENST00000569079.1 |
PRSS54
|
serine protease 54 |
| chr6_+_27808163 | 0.05 |
ENST00000358739.4
|
H2AC13
|
H2A clustered histone 13 |
| chr6_-_65707214 | 0.05 |
ENST00000370621.7
ENST00000393380.6 ENST00000503581.6 |
EYS
|
eyes shut homolog |
| chr11_-_133532493 | 0.05 |
ENST00000524381.6
|
OPCML
|
opioid binding protein/cell adhesion molecule like |
| chr11_+_61102465 | 0.05 |
ENST00000347785.8
ENST00000544014.1 |
CD5
|
CD5 molecule |
| chr18_+_32018817 | 0.04 |
ENST00000217740.4
ENST00000583184.1 |
RNF125
ENSG00000263917.1
|
ring finger protein 125 novel transcript |
| chr6_+_27133032 | 0.04 |
ENST00000359193.3
|
H2AC11
|
H2A clustered histone 11 |
| chr20_+_45408276 | 0.04 |
ENST00000372710.5
ENST00000443296.1 |
DBNDD2
|
dysbindin domain containing 2 |
| chr1_+_149832647 | 0.04 |
ENST00000578186.2
|
H4C14
|
H4 clustered histone 14 |
| chr19_+_51225059 | 0.04 |
ENST00000436584.6
ENST00000421133.6 ENST00000262262.5 ENST00000391796.7 |
CD33
|
CD33 molecule |
| chr11_+_73272201 | 0.04 |
ENST00000393590.3
|
P2RY6
|
pyrimidinergic receptor P2Y6 |
| chr14_+_91114364 | 0.04 |
ENST00000518868.5
|
DGLUCY
|
D-glutamate cyclase |
| chr14_+_91114667 | 0.04 |
ENST00000523894.5
ENST00000522322.5 ENST00000523771.5 |
DGLUCY
|
D-glutamate cyclase |
| chr6_+_27147094 | 0.04 |
ENST00000377459.3
|
H2AC12
|
H2A clustered histone 12 |
| chr14_+_91114431 | 0.04 |
ENST00000428926.6
ENST00000517362.5 |
DGLUCY
|
D-glutamate cyclase |
| chr2_+_219245455 | 0.04 |
ENST00000409638.7
ENST00000396738.7 ENST00000409516.7 |
STK16
|
serine/threonine kinase 16 |
| chr2_-_24793382 | 0.04 |
ENST00000328379.6
|
PTRHD1
|
peptidyl-tRNA hydrolase domain containing 1 |
| chr1_+_59297057 | 0.04 |
ENST00000303721.12
|
FGGY
|
FGGY carbohydrate kinase domain containing |
| chr1_+_59296971 | 0.04 |
ENST00000371218.8
|
FGGY
|
FGGY carbohydrate kinase domain containing |
| chr17_-_40937445 | 0.03 |
ENST00000436344.7
ENST00000485751.1 |
KRT23
|
keratin 23 |
| chr7_+_139829242 | 0.03 |
ENST00000455353.6
ENST00000458722.6 ENST00000448866.7 ENST00000411653.6 |
TBXAS1
|
thromboxane A synthase 1 |
| chr4_+_186069144 | 0.03 |
ENST00000513189.1
ENST00000296795.8 |
TLR3
|
toll like receptor 3 |
| chr6_-_132798587 | 0.03 |
ENST00000275227.9
|
SLC18B1
|
solute carrier family 18 member B1 |
| chr10_-_100286660 | 0.03 |
ENST00000370372.7
|
BLOC1S2
|
biogenesis of lysosomal organelles complex 1 subunit 2 |
| chr10_+_28677487 | 0.03 |
ENST00000375533.6
|
BAMBI
|
BMP and activin membrane bound inhibitor |
| chr7_-_14903319 | 0.03 |
ENST00000403951.6
|
DGKB
|
diacylglycerol kinase beta |
| chr3_-_14178569 | 0.03 |
ENST00000285021.12
|
XPC
|
XPC complex subunit, DNA damage recognition and repair factor |
| chr2_+_96325294 | 0.03 |
ENST00000439118.3
ENST00000420176.5 ENST00000536814.1 |
ITPRIPL1
|
ITPRIP like 1 |
| chr19_-_42220109 | 0.03 |
ENST00000595337.5
|
DEDD2
|
death effector domain containing 2 |
| chr19_-_45645560 | 0.03 |
ENST00000587152.6
|
EML2
|
EMAP like 2 |
| chr7_-_22822829 | 0.03 |
ENST00000358435.9
ENST00000621567.1 |
TOMM7
|
translocase of outer mitochondrial membrane 7 |
| chr6_+_47698538 | 0.03 |
ENST00000327753.7
|
ADGRF4
|
adhesion G protein-coupled receptor F4 |
| chr11_-_64744993 | 0.03 |
ENST00000377485.5
|
RASGRP2
|
RAS guanyl releasing protein 2 |
| chr6_+_47698574 | 0.03 |
ENST00000283303.3
|
ADGRF4
|
adhesion G protein-coupled receptor F4 |
| chr1_-_110607307 | 0.03 |
ENST00000639048.2
ENST00000675391.1 ENST00000639233.2 |
KCNA2
|
potassium voltage-gated channel subfamily A member 2 |
| chr6_+_27838600 | 0.03 |
ENST00000606613.1
|
H2BC15
|
H2B clustered histone 15 |
| chr6_-_26123910 | 0.03 |
ENST00000314332.5
ENST00000396984.1 |
H2BC4
|
H2B clustered histone 4 |
| chr1_-_110607425 | 0.03 |
ENST00000633222.1
|
KCNA2
|
potassium voltage-gated channel subfamily A member 2 |
| chr5_+_55851349 | 0.03 |
ENST00000652347.2
|
IL31RA
|
interleukin 31 receptor A |
| chr1_-_88891496 | 0.03 |
ENST00000448623.5
ENST00000370500.10 ENST00000418217.1 |
GTF2B
|
general transcription factor IIB |
| chr11_-_65382632 | 0.02 |
ENST00000294187.10
ENST00000398802.6 ENST00000530936.1 |
SLC25A45
|
solute carrier family 25 member 45 |
| chr7_-_22822779 | 0.02 |
ENST00000372879.8
|
TOMM7
|
translocase of outer mitochondrial membrane 7 |
| chr14_+_91114026 | 0.02 |
ENST00000521081.5
ENST00000520328.5 ENST00000524232.5 ENST00000522170.5 ENST00000256324.15 ENST00000519950.5 ENST00000523879.5 ENST00000521077.6 ENST00000518665.6 |
DGLUCY
|
D-glutamate cyclase |
| chr18_+_46946821 | 0.02 |
ENST00000245121.9
ENST00000356157.12 |
KATNAL2
|
katanin catalytic subunit A1 like 2 |
| chrX_-_27463341 | 0.02 |
ENST00000412172.4
|
PPP4R3C
|
protein phosphatase 4 regulatory subunit 3C |
| chr7_+_150405146 | 0.02 |
ENST00000498682.3
ENST00000641717.1 |
ENSG00000284691.1
|
novel zinc finger protein |
| chr19_+_41088450 | 0.02 |
ENST00000330436.4
|
CYP2A13
|
cytochrome P450 family 2 subfamily A member 13 |
| chr5_+_55851378 | 0.02 |
ENST00000396836.6
ENST00000359040.10 |
IL31RA
|
interleukin 31 receptor A |
| chr6_-_27873525 | 0.02 |
ENST00000618305.2
|
H4C13
|
H4 clustered histone 13 |
| chr3_-_154430184 | 0.02 |
ENST00000389740.3
|
GPR149
|
G protein-coupled receptor 149 |
| chr21_+_44133610 | 0.02 |
ENST00000644251.1
ENST00000427803.6 ENST00000348499.9 ENST00000291577.11 ENST00000389690.7 |
GATD3A
|
glutamine amidotransferase like class 1 domain containing 3A |
| chr18_+_13611764 | 0.02 |
ENST00000585931.5
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr11_+_117199363 | 0.02 |
ENST00000392951.9
ENST00000525531.5 ENST00000278968.10 |
TAGLN
|
transgelin |
| chr17_+_38297023 | 0.02 |
ENST00000619548.1
ENST00000613675.5 |
MRPL45
|
mitochondrial ribosomal protein L45 |
| chr7_-_19773569 | 0.02 |
ENST00000422233.5
ENST00000433641.5 ENST00000405844.6 |
TMEM196
|
transmembrane protein 196 |
| chr7_+_142656688 | 0.02 |
ENST00000390397.2
|
TRBV24-1
|
T cell receptor beta variable 24-1 |
| chr1_-_44017296 | 0.02 |
ENST00000357730.6
ENST00000360584.6 ENST00000528803.1 |
SLC6A9
|
solute carrier family 6 member 9 |
| chrX_-_154295085 | 0.02 |
ENST00000617225.4
ENST00000619903.4 |
TEX28
|
testis expressed 28 |
| chr15_+_63504583 | 0.02 |
ENST00000380324.8
ENST00000561442.5 ENST00000560070.5 |
USP3
|
ubiquitin specific peptidase 3 |
| chr15_+_63504511 | 0.02 |
ENST00000540797.5
|
USP3
|
ubiquitin specific peptidase 3 |
| chr15_+_67067780 | 0.02 |
ENST00000679624.1
|
SMAD3
|
SMAD family member 3 |
| chr6_-_26027274 | 0.02 |
ENST00000377745.4
|
H4C2
|
H4 clustered histone 2 |
| chr6_+_139028680 | 0.02 |
ENST00000367660.4
|
ABRACL
|
ABRA C-terminal like |
| chr3_+_51707059 | 0.02 |
ENST00000395052.8
|
GRM2
|
glutamate metabotropic receptor 2 |
| chr6_+_292050 | 0.02 |
ENST00000344450.9
|
DUSP22
|
dual specificity phosphatase 22 |
| chr18_+_13218195 | 0.02 |
ENST00000679167.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr15_-_82709823 | 0.02 |
ENST00000666973.1
ENST00000664460.1 ENST00000669930.1 |
AP3B2
|
adaptor related protein complex 3 subunit beta 2 |
| chr14_-_24081928 | 0.02 |
ENST00000396995.1
|
NRL
|
neural retina leucine zipper |
| chr1_-_152036984 | 0.02 |
ENST00000271638.3
|
S100A11
|
S100 calcium binding protein A11 |
| chr8_-_100105955 | 0.02 |
ENST00000523437.5
|
RGS22
|
regulator of G protein signaling 22 |
| chr14_+_91114388 | 0.02 |
ENST00000519019.5
ENST00000523816.5 ENST00000517518.5 |
DGLUCY
|
D-glutamate cyclase |
| chr1_-_9129085 | 0.01 |
ENST00000377411.5
|
GPR157
|
G protein-coupled receptor 157 |
| chr15_-_82709775 | 0.01 |
ENST00000535348.5
|
AP3B2
|
adaptor related protein complex 3 subunit beta 2 |
| chr20_-_64079479 | 0.01 |
ENST00000395042.2
|
RGS19
|
regulator of G protein signaling 19 |
| chr6_+_26272923 | 0.01 |
ENST00000377733.4
|
H2BC10
|
H2B clustered histone 10 |
| chr18_+_13612614 | 0.01 |
ENST00000586765.1
ENST00000677910.1 |
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr1_+_66332004 | 0.01 |
ENST00000371045.9
ENST00000531025.5 ENST00000526197.5 |
PDE4B
|
phosphodiesterase 4B |
| chr8_-_100106034 | 0.01 |
ENST00000360863.11
ENST00000617334.1 |
RGS22
|
regulator of G protein signaling 22 |
| chr6_+_26216928 | 0.01 |
ENST00000303910.4
|
H2AC8
|
H2A clustered histone 8 |
| chr6_+_116616467 | 0.01 |
ENST00000229554.10
ENST00000368581.8 ENST00000368580.4 |
RSPH4A
|
radial spoke head component 4A |
| chr14_+_92923143 | 0.01 |
ENST00000216492.10
ENST00000334654.4 |
CHGA
|
chromogranin A |
| chr1_-_10982037 | 0.01 |
ENST00000377004.8
|
C1orf127
|
chromosome 1 open reading frame 127 |
| chrX_-_69165430 | 0.01 |
ENST00000374584.3
ENST00000590146.1 ENST00000374571.5 |
PJA1
|
praja ring finger ubiquitin ligase 1 |
| chr16_+_2471284 | 0.01 |
ENST00000293973.2
|
NTN3
|
netrin 3 |
| chr15_-_70763430 | 0.01 |
ENST00000539319.5
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr14_-_20802402 | 0.01 |
ENST00000412779.2
|
RNASE1
|
ribonuclease A family member 1, pancreatic |
| chr9_+_133534697 | 0.01 |
ENST00000651351.2
|
ADAMTSL2
|
ADAMTS like 2 |
| chrX_-_69165509 | 0.01 |
ENST00000361478.1
|
PJA1
|
praja ring finger ubiquitin ligase 1 |
| chr16_+_30762289 | 0.01 |
ENST00000566811.5
ENST00000565995.5 ENST00000563683.5 ENST00000357890.9 ENST00000324685.11 ENST00000565931.1 |
RNF40
|
ring finger protein 40 |
| chr8_-_100105619 | 0.01 |
ENST00000523287.5
ENST00000519092.5 |
RGS22
|
regulator of G protein signaling 22 |
| chr1_-_225941383 | 0.01 |
ENST00000420304.6
|
LEFTY2
|
left-right determination factor 2 |
| chr1_-_225923891 | 0.01 |
ENST00000472798.2
ENST00000489681.5 ENST00000612651.4 |
PYCR2
|
pyrroline-5-carboxylate reductase 2 |
| chr14_-_64942783 | 0.01 |
ENST00000612794.1
|
GPX2
|
glutathione peroxidase 2 |
| chr7_+_100720463 | 0.01 |
ENST00000252723.3
|
EPO
|
erythropoietin |
| chr9_+_133534807 | 0.01 |
ENST00000393060.1
|
ADAMTSL2
|
ADAMTS like 2 |
| chr12_+_119178920 | 0.01 |
ENST00000281938.7
|
HSPB8
|
heat shock protein family B (small) member 8 |
| chr7_+_142384328 | 0.01 |
ENST00000390361.3
|
TRBV7-3
|
T cell receptor beta variable 7-3 |
| chr3_+_14178808 | 0.01 |
ENST00000306024.4
|
LSM3
|
LSM3 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr6_+_27893411 | 0.01 |
ENST00000616182.2
|
H2BC17
|
H2B clustered histone 17 |
| chr5_+_132873660 | 0.01 |
ENST00000296877.3
|
LEAP2
|
liver enriched antimicrobial peptide 2 |
| chr6_-_27838362 | 0.01 |
ENST00000618958.2
|
H2AC15
|
H2A clustered histone 15 |
| chr12_+_119178953 | 0.01 |
ENST00000674542.1
|
HSPB8
|
heat shock protein family B (small) member 8 |
| chr1_-_9069608 | 0.01 |
ENST00000377424.9
|
SLC2A5
|
solute carrier family 2 member 5 |
| chr12_+_48818763 | 0.01 |
ENST00000548279.5
ENST00000547230.5 |
CACNB3
|
calcium voltage-gated channel auxiliary subunit beta 3 |
| chr21_+_31659666 | 0.01 |
ENST00000389995.4
ENST00000270142.11 |
SOD1
|
superoxide dismutase 1 |
| chr14_-_24081986 | 0.01 |
ENST00000560550.1
|
NRL
|
neural retina leucine zipper |
| chr3_+_125969152 | 0.01 |
ENST00000251776.8
ENST00000504401.1 |
ROPN1B
|
rhophilin associated tail protein 1B |
| chr4_+_2812259 | 0.01 |
ENST00000502260.5
ENST00000435136.8 |
SH3BP2
|
SH3 domain binding protein 2 |
| chr13_+_45464901 | 0.01 |
ENST00000349995.10
|
COG3
|
component of oligomeric golgi complex 3 |
| chr6_-_53148822 | 0.01 |
ENST00000259803.8
|
GCM1
|
glial cells missing transcription factor 1 |
| chr1_-_225941212 | 0.01 |
ENST00000366820.10
|
LEFTY2
|
left-right determination factor 2 |
| chr7_+_50308672 | 0.01 |
ENST00000439701.2
ENST00000438033.5 ENST00000492782.6 |
IKZF1
|
IKAROS family zinc finger 1 |
| chr15_+_51341648 | 0.01 |
ENST00000335449.11
ENST00000560215.5 |
GLDN
|
gliomedin |
| chr20_-_6123019 | 0.01 |
ENST00000217289.9
ENST00000536936.1 |
FERMT1
|
fermitin family member 1 |
| chr14_-_64942720 | 0.01 |
ENST00000557049.1
ENST00000389614.6 |
GPX2
|
glutathione peroxidase 2 |
| chr15_-_82806054 | 0.01 |
ENST00000541889.1
ENST00000334574.12 ENST00000561368.1 |
FSD2
|
fibronectin type III and SPRY domain containing 2 |
| chr19_+_16077006 | 0.01 |
ENST00000586833.7
ENST00000642221.1 |
TPM4
|
tropomyosin 4 |
| chr12_+_14774362 | 0.01 |
ENST00000544848.2
|
H2AJ
|
H2A.J histone |
| chr14_-_58152179 | 0.01 |
ENST00000267485.7
|
ARMH4
|
armadillo like helical domain containing 4 |
| chr3_-_112846856 | 0.01 |
ENST00000488794.5
|
CD200R1L
|
CD200 receptor 1 like |
| chr5_+_141245384 | 0.01 |
ENST00000623671.1
ENST00000231173.6 |
PCDHB15
|
protocadherin beta 15 |
| chr16_-_20669855 | 0.01 |
ENST00000524149.5
|
ACSM1
|
acyl-CoA synthetase medium chain family member 1 |
| chr17_+_4710622 | 0.01 |
ENST00000574954.5
ENST00000269260.7 ENST00000346341.6 ENST00000572457.5 ENST00000381488.10 ENST00000412477.7 ENST00000571428.5 ENST00000575877.5 |
ARRB2
|
arrestin beta 2 |
| chr4_-_147684114 | 0.00 |
ENST00000322396.7
|
PRMT9
|
protein arginine methyltransferase 9 |
| chr1_-_9069572 | 0.00 |
ENST00000377414.7
|
SLC2A5
|
solute carrier family 2 member 5 |
| chr15_+_82262781 | 0.00 |
ENST00000566205.1
ENST00000339465.5 ENST00000569120.1 ENST00000682753.1 ENST00000566861.5 ENST00000565432.1 |
SAXO2
|
stabilizer of axonemal microtubules 2 |
| chr16_+_30472733 | 0.00 |
ENST00000356798.11
ENST00000433423.2 |
ITGAL
|
integrin subunit alpha L |
| chr16_-_30762052 | 0.00 |
ENST00000543610.6
ENST00000545825.1 ENST00000541260.5 |
CCDC189
|
coiled-coil domain containing 189 |
| chr1_+_202416826 | 0.00 |
ENST00000466968.1
|
PPP1R12B
|
protein phosphatase 1 regulatory subunit 12B |
| chr8_-_118951876 | 0.00 |
ENST00000297350.9
|
TNFRSF11B
|
TNF receptor superfamily member 11b |
| chr9_-_34691204 | 0.00 |
ENST00000378800.3
ENST00000311925.7 |
CCL19
|
C-C motif chemokine ligand 19 |
| chr10_+_127998126 | 0.00 |
ENST00000455661.5
|
PTPRE
|
protein tyrosine phosphatase receptor type E |
| chr11_-_72434330 | 0.00 |
ENST00000542555.2
ENST00000535990.5 ENST00000294053.9 ENST00000538039.6 ENST00000340729.9 |
CLPB
|
caseinolytic mitochondrial matrix peptidase chaperone subunit B |
| chr3_-_120647018 | 0.00 |
ENST00000475447.2
|
HGD
|
homogentisate 1,2-dioxygenase |
| chr5_-_69332723 | 0.00 |
ENST00000511257.1
ENST00000396496.7 ENST00000383374.6 |
CCDC125
|
coiled-coil domain containing 125 |
| chr10_+_69278492 | 0.00 |
ENST00000643399.2
|
HK1
|
hexokinase 1 |
| chr11_+_66638678 | 0.00 |
ENST00000578778.5
ENST00000483858.5 ENST00000398692.8 ENST00000310092.12 ENST00000510173.6 ENST00000506523.6 ENST00000530235.1 ENST00000532968.1 |
RBM4
|
RNA binding motif protein 4 |
| chr5_-_133026533 | 0.00 |
ENST00000509437.6
ENST00000355372.6 ENST00000513541.5 ENST00000509008.1 ENST00000513848.5 ENST00000504170.2 ENST00000324170.7 |
ZCCHC10
|
zinc finger CCHC-type containing 10 |
| chr1_-_60073750 | 0.00 |
ENST00000371201.3
|
C1orf87
|
chromosome 1 open reading frame 87 |
| chr16_-_84186998 | 0.00 |
ENST00000567759.5
|
TAF1C
|
TATA-box binding protein associated factor, RNA polymerase I subunit C |
| chrX_-_49073989 | 0.00 |
ENST00000376386.3
ENST00000553851.3 |
PRAF2
|
PRA1 domain family member 2 |
| chr1_-_60073733 | 0.00 |
ENST00000450089.6
|
C1orf87
|
chromosome 1 open reading frame 87 |
| chr1_-_36440873 | 0.00 |
ENST00000433045.6
|
OSCP1
|
organic solute carrier partner 1 |
| chr6_-_26285526 | 0.00 |
ENST00000377727.2
|
H4C8
|
H4 clustered histone 8 |
| chr1_-_160647287 | 0.00 |
ENST00000235739.6
|
SLAMF1
|
signaling lymphocytic activation molecule family member 1 |
| chr12_+_67269328 | 0.00 |
ENST00000545606.6
|
CAND1
|
cullin associated and neddylation dissociated 1 |
| chr19_+_42220283 | 0.00 |
ENST00000301215.8
ENST00000597945.1 |
ZNF526
|
zinc finger protein 526 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SMAD3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0051945 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.0 | 0.1 | GO:0061011 | hepatic duct development(GO:0061011) |
| 0.0 | 0.1 | GO:0072144 | glomerular mesangial cell development(GO:0072144) mesenchyme migration(GO:0090131) |
| 0.0 | 0.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0010265 | SCF complex assembly(GO:0010265) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.2 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.1 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.0 | 0.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.0 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |