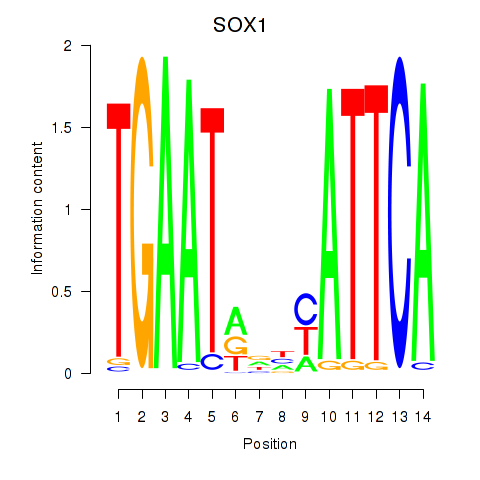
|
chr6_+_130421086
Show fit
|
0.57 |
ENST00000545622.5
|
TMEM200A
|
transmembrane protein 200A
|
|
chr3_-_190122317
Show fit
|
0.56 |
ENST00000427335.6
|
P3H2
|
prolyl 3-hydroxylase 2
|
|
chr16_+_11965193
Show fit
|
0.39 |
ENST00000053243.6
ENST00000396495.3
|
TNFRSF17
|
TNF receptor superfamily member 17
|
|
chr3_+_136957948
Show fit
|
0.34 |
ENST00000329582.9
|
IL20RB
|
interleukin 20 receptor subunit beta
|
|
chr1_+_152985231
Show fit
|
0.28 |
ENST00000368762.1
|
SPRR1A
|
small proline rich protein 1A
|
|
chr1_-_12616762
Show fit
|
0.27 |
ENST00000464917.5
|
DHRS3
|
dehydrogenase/reductase 3
|
|
chr2_+_101998955
Show fit
|
0.26 |
ENST00000393414.6
|
IL1R2
|
interleukin 1 receptor type 2
|
|
chr8_-_109680812
Show fit
|
0.24 |
ENST00000528716.5
ENST00000527600.5
ENST00000531230.5
ENST00000532189.5
ENST00000534184.5
ENST00000408889.7
ENST00000533171.5
|
SYBU
|
syntabulin
|
|
chr1_+_70411241
Show fit
|
0.20 |
ENST00000370938.8
ENST00000346806.2
|
CTH
|
cystathionine gamma-lyase
|
|
chr8_-_109608055
Show fit
|
0.19 |
ENST00000529690.5
|
SYBU
|
syntabulin
|
|
chr13_+_108596152
Show fit
|
0.19 |
ENST00000356711.7
ENST00000251041.10
|
MYO16
|
myosin XVI
|
|
chr17_-_41489907
Show fit
|
0.18 |
ENST00000328119.11
|
KRT36
|
keratin 36
|
|
chr13_+_32316063
Show fit
|
0.18 |
ENST00000680887.1
|
BRCA2
|
BRCA2 DNA repair associated
|
|
chr20_-_14337602
Show fit
|
0.18 |
ENST00000378053.3
ENST00000341420.5
|
FLRT3
|
fibronectin leucine rich transmembrane protein 3
|
|
chrX_+_106168297
Show fit
|
0.16 |
ENST00000337685.6
ENST00000357175.6
|
PWWP3B
|
PWWP domain containing 3B
|
|
chr11_-_133845495
Show fit
|
0.13 |
ENST00000299140.8
ENST00000532889.1
|
SPATA19
|
spermatogenesis associated 19
|
|
chr1_+_32209074
Show fit
|
0.12 |
ENST00000409358.2
|
DCDC2B
|
doublecortin domain containing 2B
|
|
chr9_-_101487120
Show fit
|
0.12 |
ENST00000374848.8
|
PGAP4
|
post-GPI attachment to proteins GalNAc transferase 4
|
|
chr4_-_73620629
Show fit
|
0.12 |
ENST00000342081.7
|
RASSF6
|
Ras association domain family member 6
|
|
chr8_+_144078590
Show fit
|
0.12 |
ENST00000525936.1
|
EXOSC4
|
exosome component 4
|
|
chr5_-_16916400
Show fit
|
0.12 |
ENST00000513882.5
|
MYO10
|
myosin X
|
|
chr17_-_31297231
Show fit
|
0.11 |
ENST00000247271.5
|
OMG
|
oligodendrocyte myelin glycoprotein
|
|
chr8_+_144078661
Show fit
|
0.11 |
ENST00000316052.6
|
EXOSC4
|
exosome component 4
|
|
chr9_-_2844058
Show fit
|
0.11 |
ENST00000397885.3
|
PUM3
|
pumilio RNA binding family member 3
|
|
chr12_-_120325936
Show fit
|
0.11 |
ENST00000549767.1
|
PLA2G1B
|
phospholipase A2 group IB
|
|
chrX_-_32412220
Show fit
|
0.11 |
ENST00000619831.5
|
DMD
|
dystrophin
|
|
chr3_-_33645433
Show fit
|
0.11 |
ENST00000635664.1
ENST00000485378.6
ENST00000313350.10
ENST00000487200.5
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr1_-_200410001
Show fit
|
0.10 |
ENST00000367353.2
|
ZNF281
|
zinc finger protein 281
|
|
chr4_+_69995958
Show fit
|
0.10 |
ENST00000381060.2
ENST00000246895.9
|
STATH
|
statherin
|
|
chr11_-_57410113
Show fit
|
0.09 |
ENST00000529411.1
|
ENSG00000254979.5
|
novel protein
|
|
chrX_+_129906146
Show fit
|
0.09 |
ENST00000394422.8
|
UTP14A
|
UTP14A small subunit processome component
|
|
chr6_+_26183750
Show fit
|
0.08 |
ENST00000614097.3
|
H2BC6
|
H2B clustered histone 6
|
|
chr17_-_69150062
Show fit
|
0.08 |
ENST00000522787.5
ENST00000521538.5
|
ABCA10
|
ATP binding cassette subfamily A member 10
|
|
chr6_-_70957029
Show fit
|
0.08 |
ENST00000230053.11
|
B3GAT2
|
beta-1,3-glucuronyltransferase 2
|
|
chr3_-_58214671
Show fit
|
0.08 |
ENST00000460422.1
ENST00000483681.5
|
DNASE1L3
|
deoxyribonuclease 1 like 3
|
|
chr2_-_162318129
Show fit
|
0.08 |
ENST00000679938.1
|
IFIH1
|
interferon induced with helicase C domain 1
|
|
chrX_+_129906118
Show fit
|
0.07 |
ENST00000425117.6
|
UTP14A
|
UTP14A small subunit processome component
|
|
chr1_-_200409976
Show fit
|
0.07 |
ENST00000367352.3
|
ZNF281
|
zinc finger protein 281
|
|
chr11_+_6863057
Show fit
|
0.07 |
ENST00000641461.1
|
OR10A2
|
olfactory receptor family 10 subfamily A member 2
|
|
chr3_+_158644510
Show fit
|
0.07 |
ENST00000486715.6
ENST00000478576.5
ENST00000264263.9
ENST00000464732.1
|
GFM1
|
G elongation factor mitochondrial 1
|
|
chr4_-_74794514
Show fit
|
0.07 |
ENST00000395743.8
|
BTC
|
betacellulin
|
|
chr11_+_4704782
Show fit
|
0.07 |
ENST00000380390.6
|
MMP26
|
matrix metallopeptidase 26
|
|
chr1_-_200410052
Show fit
|
0.07 |
ENST00000294740.3
|
ZNF281
|
zinc finger protein 281
|
|
chr12_-_9999176
Show fit
|
0.07 |
ENST00000298527.10
ENST00000348658.4
|
CLEC1B
|
C-type lectin domain family 1 member B
|
|
chr2_-_208190001
Show fit
|
0.06 |
ENST00000451346.5
ENST00000341287.9
|
C2orf80
|
chromosome 2 open reading frame 80
|
|
chr1_-_169485931
Show fit
|
0.06 |
ENST00000367804.4
ENST00000646596.1
ENST00000236137.10
|
SLC19A2
|
solute carrier family 19 member 2
|
|
chr19_+_17719471
Show fit
|
0.06 |
ENST00000600186.5
ENST00000597735.5
ENST00000324096.9
|
MAP1S
|
microtubule associated protein 1S
|
|
chr11_-_47378494
Show fit
|
0.06 |
ENST00000533030.1
|
SPI1
|
Spi-1 proto-oncogene
|
|
chr6_+_123804141
Show fit
|
0.06 |
ENST00000368416.5
|
NKAIN2
|
sodium/potassium transporting ATPase interacting 2
|
|
chr9_-_127847117
Show fit
|
0.06 |
ENST00000480266.5
|
ENG
|
endoglin
|
|
chr11_-_47378391
Show fit
|
0.06 |
ENST00000227163.8
|
SPI1
|
Spi-1 proto-oncogene
|
|
chr1_+_174799895
Show fit
|
0.06 |
ENST00000489615.5
|
RABGAP1L
|
RAB GTPase activating protein 1 like
|
|
chrX_-_72877864
Show fit
|
0.06 |
ENST00000596389.5
|
DMRTC1
|
DMRT like family C1
|
|
chr1_-_183653307
Show fit
|
0.06 |
ENST00000308641.6
|
APOBEC4
|
apolipoprotein B mRNA editing enzyme catalytic polypeptide like 4
|
|
chr20_+_33217325
Show fit
|
0.05 |
ENST00000375452.3
ENST00000375454.8
|
BPIFA3
|
BPI fold containing family A member 3
|
|
chr10_-_43397220
Show fit
|
0.05 |
ENST00000477108.5
ENST00000544000.5
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F
|
|
chr1_-_43172504
Show fit
|
0.05 |
ENST00000431635.6
|
EBNA1BP2
|
EBNA1 binding protein 2
|
|
chr1_-_19251509
Show fit
|
0.05 |
ENST00000375199.7
ENST00000375208.7
ENST00000477853.6
|
EMC1
|
ER membrane protein complex subunit 1
|
|
chr17_+_36064265
Show fit
|
0.04 |
ENST00000616054.2
|
CCL18
|
C-C motif chemokine ligand 18
|
|
chr19_+_35351552
Show fit
|
0.04 |
ENST00000246553.3
|
FFAR1
|
free fatty acid receptor 1
|
|
chr11_+_103109398
Show fit
|
0.04 |
ENST00000648198.1
ENST00000375735.7
ENST00000650373.2
|
DYNC2H1
|
dynein cytoplasmic 2 heavy chain 1
|
|
chr2_-_162318475
Show fit
|
0.04 |
ENST00000648433.1
|
IFIH1
|
interferon induced with helicase C domain 1
|
|
chr13_-_60163764
Show fit
|
0.04 |
ENST00000377908.6
ENST00000400319.5
ENST00000400320.5
ENST00000267215.8
|
DIAPH3
|
diaphanous related formin 3
|
|
chr10_+_76318330
Show fit
|
0.04 |
ENST00000496424.2
|
LRMDA
|
leucine rich melanocyte differentiation associated
|
|
chr1_-_52033772
Show fit
|
0.04 |
ENST00000371614.2
|
KTI12
|
KTI12 chromatin associated homolog
|
|
chr19_+_49085433
Show fit
|
0.04 |
ENST00000221448.9
ENST00000598441.6
|
SNRNP70
|
small nuclear ribonucleoprotein U1 subunit 70
|
|
chr11_-_113706330
Show fit
|
0.04 |
ENST00000540540.5
ENST00000538955.5
ENST00000545579.6
|
TMPRSS5
|
transmembrane serine protease 5
|
|
chr15_+_40382715
Show fit
|
0.04 |
ENST00000416151.6
ENST00000249776.12
|
KNSTRN
|
kinetochore localized astrin (SPAG5) binding protein
|
|
chr22_-_30266839
Show fit
|
0.03 |
ENST00000403463.1
ENST00000215781.3
|
OSM
|
oncostatin M
|
|
chr4_-_170003738
Show fit
|
0.03 |
ENST00000502832.1
ENST00000393704.3
|
MFAP3L
|
microfibril associated protein 3 like
|
|
chr2_+_170178136
Show fit
|
0.03 |
ENST00000409044.7
ENST00000408978.9
|
MYO3B
|
myosin IIIB
|
|
chr3_+_178558700
Show fit
|
0.03 |
ENST00000432997.5
ENST00000455865.5
|
KCNMB2
|
potassium calcium-activated channel subfamily M regulatory beta subunit 2
|
|
chr15_-_45114149
Show fit
|
0.03 |
ENST00000603300.1
ENST00000389039.11
|
DUOX2
|
dual oxidase 2
|
|
chr10_-_72217126
Show fit
|
0.03 |
ENST00000317126.8
|
ASCC1
|
activating signal cointegrator 1 complex subunit 1
|
|
chr15_+_59611776
Show fit
|
0.03 |
ENST00000396065.3
ENST00000560585.5
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type
|
|
chr6_+_63635792
Show fit
|
0.03 |
ENST00000262043.8
ENST00000506783.5
ENST00000481385.6
ENST00000515594.5
ENST00000494284.6
|
PHF3
|
PHD finger protein 3
|
|
chr7_-_105389365
Show fit
|
0.03 |
ENST00000482897.5
|
SRPK2
|
SRSF protein kinase 2
|
|
chr7_-_26200734
Show fit
|
0.03 |
ENST00000354667.8
ENST00000618183.5
|
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1
|
|
chr17_-_17281232
Show fit
|
0.03 |
ENST00000417352.5
ENST00000268717.10
|
COPS3
|
COP9 signalosome subunit 3
|
|
chr8_-_85378105
Show fit
|
0.02 |
ENST00000521846.5
ENST00000523022.6
ENST00000524324.5
ENST00000519991.5
ENST00000520663.5
ENST00000517590.5
ENST00000522579.5
ENST00000522814.5
ENST00000522662.5
ENST00000523858.5
ENST00000519129.5
|
CA1
|
carbonic anhydrase 1
|
|
chr7_-_93219564
Show fit
|
0.02 |
ENST00000341723.8
|
HEPACAM2
|
HEPACAM family member 2
|
|
chr7_-_93219545
Show fit
|
0.02 |
ENST00000440868.5
|
HEPACAM2
|
HEPACAM family member 2
|
|
chr3_+_128051610
Show fit
|
0.02 |
ENST00000464451.5
|
SEC61A1
|
SEC61 translocon subunit alpha 1
|
|
chrX_+_49171889
Show fit
|
0.02 |
ENST00000376327.6
|
PLP2
|
proteolipid protein 2
|
|
chr11_-_113706292
Show fit
|
0.02 |
ENST00000544634.5
ENST00000539732.5
ENST00000538770.1
ENST00000536856.5
ENST00000299882.11
ENST00000544476.1
|
TMPRSS5
|
transmembrane serine protease 5
|
|
chr15_-_73753308
Show fit
|
0.02 |
ENST00000569673.3
|
INSYN1
|
inhibitory synaptic factor 1
|
|
chr19_-_10380454
Show fit
|
0.02 |
ENST00000530829.1
ENST00000529370.5
|
TYK2
|
tyrosine kinase 2
|
|
chr7_-_86965872
Show fit
|
0.02 |
ENST00000398276.6
ENST00000416314.5
ENST00000425689.1
|
ELAPOR2
|
endosome-lysosome associated apoptosis and autophagy regulator family member 2
|
|
chr1_+_240091866
Show fit
|
0.02 |
ENST00000319653.14
|
FMN2
|
formin 2
|
|
chr1_+_84479239
Show fit
|
0.02 |
ENST00000370656.5
ENST00000370654.6
|
RPF1
|
ribosome production factor 1 homolog
|
|
chr10_-_72216267
Show fit
|
0.02 |
ENST00000342444.8
ENST00000533958.1
ENST00000672957.1
ENST00000527593.5
ENST00000672940.1
ENST00000530461.5
ENST00000317168.11
ENST00000524829.5
|
ASCC1
|
activating signal cointegrator 1 complex subunit 1
|
|
chr18_+_24139053
Show fit
|
0.02 |
ENST00000463087.5
ENST00000585037.5
ENST00000399496.8
ENST00000486759.6
ENST00000577705.1
ENST00000415309.6
ENST00000621648.4
ENST00000581397.5
|
CABYR
|
calcium binding tyrosine phosphorylation regulated
|
|
chr9_-_111328520
Show fit
|
0.02 |
ENST00000374428.1
|
OR2K2
|
olfactory receptor family 2 subfamily K member 2
|
|
chr17_-_4641670
Show fit
|
0.02 |
ENST00000293761.8
|
ALOX15
|
arachidonate 15-lipoxygenase
|
|
chr1_-_86383078
Show fit
|
0.02 |
ENST00000460698.6
|
ODF2L
|
outer dense fiber of sperm tails 2 like
|
|
chr2_-_162318613
Show fit
|
0.02 |
ENST00000649979.2
ENST00000421365.2
|
IFIH1
|
interferon induced with helicase C domain 1
|
|
chr18_-_54224578
Show fit
|
0.02 |
ENST00000583046.1
ENST00000398398.6
ENST00000256429.8
|
MBD2
|
methyl-CpG binding domain protein 2
|
|
chr3_+_11011640
Show fit
|
0.02 |
ENST00000643396.1
|
SLC6A1
|
solute carrier family 6 member 1
|
|
chr19_+_51761167
Show fit
|
0.02 |
ENST00000340023.7
ENST00000599326.1
ENST00000598953.1
|
FPR2
|
formyl peptide receptor 2
|
|
chr17_+_77185210
Show fit
|
0.02 |
ENST00000431431.6
|
SEC14L1
|
SEC14 like lipid binding 1
|
|
chr17_-_40665121
Show fit
|
0.01 |
ENST00000394052.5
|
KRT222
|
keratin 222
|
|
chr11_-_26572130
Show fit
|
0.01 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated
|
|
chrX_-_56995508
Show fit
|
0.01 |
ENST00000374919.6
ENST00000639007.1
ENST00000639583.1
ENST00000638289.1
ENST00000639525.1
ENST00000638386.1
|
SPIN3
|
spindlin family member 3
|
|
chr11_+_125626229
Show fit
|
0.01 |
ENST00000532449.6
ENST00000534070.5
|
CHEK1
|
checkpoint kinase 1
|
|
chr1_+_65147514
Show fit
|
0.01 |
ENST00000545314.5
|
AK4
|
adenylate kinase 4
|
|
chrX_-_56995426
Show fit
|
0.01 |
ENST00000640768.1
ENST00000638619.1
|
SPIN3
|
spindlin family member 3
|
|
chr1_+_203795614
Show fit
|
0.01 |
ENST00000367210.3
ENST00000432282.5
ENST00000453771.5
ENST00000367214.5
ENST00000639812.1
ENST00000367212.7
ENST00000332127.8
ENST00000550078.2
|
ZC3H11A
ZBED6
|
zinc finger CCCH-type containing 11A
zinc finger BED-type containing 6
|
|
chr12_-_109996291
Show fit
|
0.01 |
ENST00000320063.10
ENST00000457474.6
ENST00000547815.5
ENST00000361006.9
ENST00000355312.8
ENST00000354574.8
ENST00000553118.5
|
GIT2
|
GIT ArfGAP 2
|
|
chr4_+_155758990
Show fit
|
0.00 |
ENST00000505154.5
ENST00000652626.1
ENST00000502959.5
ENST00000264424.13
ENST00000505764.5
ENST00000507146.5
ENST00000503520.5
|
GUCY1B1
|
guanylate cyclase 1 soluble subunit beta 1
|
|
chr12_+_92702843
Show fit
|
0.00 |
ENST00000397833.3
|
PLEKHG7
|
pleckstrin homology and RhoGEF domain containing G7
|
|
chr14_-_60165363
Show fit
|
0.00 |
ENST00000557185.6
|
DHRS7
|
dehydrogenase/reductase 7
|
|
chr19_+_54874218
Show fit
|
0.00 |
ENST00000355524.8
ENST00000391726.7
|
FCAR
|
Fc fragment of IgA receptor
|
|
chrX_+_11111291
Show fit
|
0.00 |
ENST00000321143.8
ENST00000380762.5
ENST00000380763.7
|
HCCS
|
holocytochrome c synthase
|