Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for SOX17
Z-value: 0.63
Transcription factors associated with SOX17
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX17
|
ENSG00000164736.6 | SRY-box transcription factor 17 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX17 | hg38_v1_chr8_+_54457927_54457943 | -0.33 | 4.3e-01 | Click! |
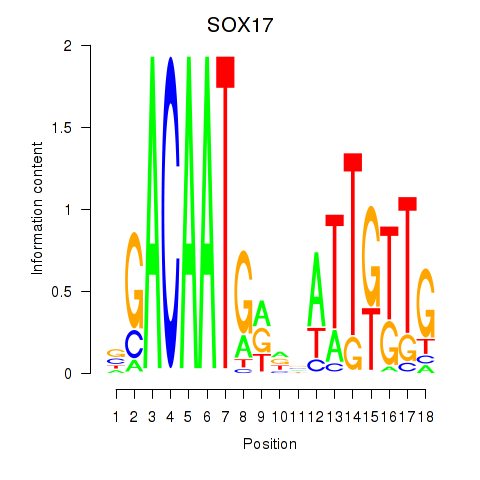
Activity profile of SOX17 motif
Sorted Z-values of SOX17 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_125153649 | 0.55 |
ENST00000304877.17
ENST00000368402.9 ENST00000368388.6 ENST00000534000.6 |
TPD52L1
|
TPD52 like 1 |
| chr8_+_94641199 | 0.53 |
ENST00000646773.1
ENST00000454170.7 |
ESRP1
|
epithelial splicing regulatory protein 1 |
| chr12_-_8662703 | 0.47 |
ENST00000535336.5
|
MFAP5
|
microfibril associated protein 5 |
| chr12_-_8662073 | 0.46 |
ENST00000535411.5
ENST00000540087.5 |
MFAP5
|
microfibril associated protein 5 |
| chr12_-_8662619 | 0.45 |
ENST00000544889.1
ENST00000543369.5 |
MFAP5
|
microfibril associated protein 5 |
| chr1_+_14929734 | 0.44 |
ENST00000376028.8
ENST00000400798.6 |
KAZN
|
kazrin, periplakin interacting protein |
| chr11_-_125496122 | 0.41 |
ENST00000527534.1
ENST00000278919.8 ENST00000366139.3 |
FEZ1
|
fasciculation and elongation protein zeta 1 |
| chr2_-_215138603 | 0.41 |
ENST00000272895.12
|
ABCA12
|
ATP binding cassette subfamily A member 12 |
| chr7_+_148339452 | 0.41 |
ENST00000463592.3
|
CNTNAP2
|
contactin associated protein 2 |
| chr19_+_6135635 | 0.39 |
ENST00000588304.5
ENST00000588722.5 ENST00000588485.6 ENST00000591403.5 ENST00000586696.5 ENST00000681525.1 ENST00000589401.5 |
ACSBG2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr11_-_118252279 | 0.38 |
ENST00000525386.5
ENST00000527472.1 ENST00000278949.9 |
MPZL3
|
myelin protein zero like 3 |
| chr1_+_43530847 | 0.36 |
ENST00000617451.4
ENST00000359947.9 ENST00000438120.5 |
PTPRF
|
protein tyrosine phosphatase receptor type F |
| chr17_-_76532046 | 0.32 |
ENST00000590175.5
|
CYGB
|
cytoglobin |
| chr12_-_8650529 | 0.32 |
ENST00000543467.5
|
MFAP5
|
microfibril associated protein 5 |
| chr14_+_21057822 | 0.31 |
ENST00000308227.2
|
RNASE8
|
ribonuclease A family member 8 |
| chr18_+_63775395 | 0.29 |
ENST00000398019.7
|
SERPINB7
|
serpin family B member 7 |
| chr22_-_28711931 | 0.28 |
ENST00000434810.5
ENST00000456369.5 |
CHEK2
|
checkpoint kinase 2 |
| chr7_-_22194709 | 0.28 |
ENST00000458533.5
|
RAPGEF5
|
Rap guanine nucleotide exchange factor 5 |
| chr19_+_14941489 | 0.28 |
ENST00000248072.3
|
OR7C2
|
olfactory receptor family 7 subfamily C member 2 |
| chr22_-_28712136 | 0.28 |
ENST00000464581.6
|
CHEK2
|
checkpoint kinase 2 |
| chr1_+_84408230 | 0.27 |
ENST00000370662.3
|
DNASE2B
|
deoxyribonuclease 2 beta |
| chr5_-_178187364 | 0.25 |
ENST00000463439.3
|
GMCL2
|
germ cell-less 2, spermatogenesis associated |
| chr6_+_30888672 | 0.24 |
ENST00000446312.5
|
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr11_-_6030758 | 0.23 |
ENST00000641900.1
|
OR56A1
|
olfactory receptor family 56 subfamily A member 1 |
| chr16_+_2817230 | 0.23 |
ENST00000005995.8
ENST00000574813.5 |
PRSS21
|
serine protease 21 |
| chr1_+_172452885 | 0.23 |
ENST00000367725.4
|
C1orf105
|
chromosome 1 open reading frame 105 |
| chr8_+_22059198 | 0.22 |
ENST00000523266.5
ENST00000519907.5 |
DMTN
|
dematin actin binding protein |
| chr6_-_132659178 | 0.22 |
ENST00000275216.3
|
TAAR1
|
trace amine associated receptor 1 |
| chr1_+_75796867 | 0.22 |
ENST00000263187.4
|
MSH4
|
mutS homolog 4 |
| chr8_+_22059342 | 0.20 |
ENST00000415253.5
|
DMTN
|
dematin actin binding protein |
| chr12_-_8662808 | 0.20 |
ENST00000359478.7
ENST00000396549.6 |
MFAP5
|
microfibril associated protein 5 |
| chr3_+_46370854 | 0.20 |
ENST00000292303.4
|
CCR5
|
C-C motif chemokine receptor 5 |
| chr5_+_55102635 | 0.19 |
ENST00000274306.7
|
GZMA
|
granzyme A |
| chr22_+_44069043 | 0.19 |
ENST00000404989.1
|
PARVB
|
parvin beta |
| chr6_+_29395631 | 0.19 |
ENST00000642051.1
|
OR12D2
|
olfactory receptor family 12 subfamily D member 2 |
| chr17_+_59220446 | 0.19 |
ENST00000284116.9
ENST00000581140.5 ENST00000581276.5 |
GDPD1
|
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr3_+_111542178 | 0.19 |
ENST00000283285.10
ENST00000352690.9 |
CD96
|
CD96 molecule |
| chr11_+_59755365 | 0.19 |
ENST00000337979.9
|
STX3
|
syntaxin 3 |
| chr14_+_94110728 | 0.19 |
ENST00000616764.4
ENST00000618863.1 ENST00000611954.4 ENST00000618200.4 ENST00000621160.4 ENST00000555819.5 ENST00000620396.4 ENST00000612813.4 ENST00000620066.1 |
IFI27
|
interferon alpha inducible protein 27 |
| chr7_+_124476371 | 0.18 |
ENST00000473520.1
|
SSU72P8
|
SSU72 pseudogene 8 |
| chr14_-_70416973 | 0.18 |
ENST00000555276.5
ENST00000617124.4 |
COX16
SYNJ2BP-COX16
|
cytochrome c oxidase assembly factor COX16 SYNJ2BP-COX16 readthrough |
| chr11_+_4233288 | 0.17 |
ENST00000639584.1
|
SSU72P5
|
SSU72 pseudogene 5 |
| chr2_+_181457342 | 0.17 |
ENST00000397033.7
ENST00000233573.6 |
ITGA4
|
integrin subunit alpha 4 |
| chr19_+_34404379 | 0.17 |
ENST00000246535.4
|
PDCD2L
|
programmed cell death 2 like |
| chr19_-_53267723 | 0.16 |
ENST00000311170.5
|
VN1R4
|
vomeronasal 1 receptor 4 |
| chr2_-_89222461 | 0.16 |
ENST00000482769.1
|
IGKV2-28
|
immunoglobulin kappa variable 2-28 |
| chr6_-_142147122 | 0.16 |
ENST00000258042.2
|
NMBR
|
neuromedin B receptor |
| chr1_+_22007450 | 0.16 |
ENST00000400271.2
|
CELA3A
|
chymotrypsin like elastase 3A |
| chr18_+_58221535 | 0.15 |
ENST00000431212.6
ENST00000586268.5 ENST00000587190.5 |
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr16_+_3654683 | 0.15 |
ENST00000246949.10
|
DNASE1
|
deoxyribonuclease 1 |
| chr12_+_22625357 | 0.15 |
ENST00000545979.2
|
ETNK1
|
ethanolamine kinase 1 |
| chr14_+_39233908 | 0.15 |
ENST00000280082.4
|
MIA2
|
MIA SH3 domain ER export factor 2 |
| chr22_-_18936142 | 0.15 |
ENST00000438924.5
ENST00000457083.1 ENST00000357068.11 ENST00000420436.5 ENST00000334029.6 ENST00000610940.4 |
PRODH
|
proline dehydrogenase 1 |
| chrX_-_13319952 | 0.14 |
ENST00000622204.1
ENST00000380622.5 |
ATXN3L
|
ataxin 3 like |
| chr1_-_109393197 | 0.14 |
ENST00000538502.5
ENST00000482236.5 |
SORT1
|
sortilin 1 |
| chr11_+_57805541 | 0.14 |
ENST00000683201.1
ENST00000683769.1 |
CTNND1
|
catenin delta 1 |
| chr2_+_233718734 | 0.13 |
ENST00000373409.8
|
UGT1A4
|
UDP glucuronosyltransferase family 1 member A4 |
| chr1_-_26360050 | 0.13 |
ENST00000475866.3
|
CRYBG2
|
crystallin beta-gamma domain containing 2 |
| chr11_-_44950867 | 0.13 |
ENST00000528290.5
ENST00000525680.6 ENST00000530035.5 ENST00000527685.5 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr2_+_102418642 | 0.13 |
ENST00000264260.6
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chr4_-_149815826 | 0.13 |
ENST00000636793.2
ENST00000636414.1 |
IQCM
|
IQ motif containing M |
| chr3_+_125969172 | 0.12 |
ENST00000514116.6
ENST00000513830.5 |
ROPN1B
|
rhophilin associated tail protein 1B |
| chr12_+_69825221 | 0.12 |
ENST00000552032.7
|
MYRFL
|
myelin regulatory factor like |
| chr11_-_123885627 | 0.12 |
ENST00000528595.1
ENST00000375026.7 |
TMEM225
|
transmembrane protein 225 |
| chr19_+_2163915 | 0.12 |
ENST00000398665.8
|
DOT1L
|
DOT1 like histone lysine methyltransferase |
| chr1_-_160523204 | 0.12 |
ENST00000368055.1
ENST00000368057.8 ENST00000368059.7 |
SLAMF6
|
SLAM family member 6 |
| chr21_+_43169008 | 0.12 |
ENST00000291554.6
|
CRYAA
|
crystallin alpha A |
| chr7_-_24757926 | 0.12 |
ENST00000342947.9
ENST00000419307.6 |
GSDME
|
gasdermin E |
| chr2_+_90172802 | 0.11 |
ENST00000390277.3
|
IGKV3D-11
|
immunoglobulin kappa variable 3D-11 |
| chr11_+_60396435 | 0.11 |
ENST00000395005.6
|
MS4A14
|
membrane spanning 4-domains A14 |
| chr12_+_8992029 | 0.11 |
ENST00000543895.1
|
KLRG1
|
killer cell lectin like receptor G1 |
| chr12_+_69825273 | 0.11 |
ENST00000547771.6
|
MYRFL
|
myelin regulatory factor like |
| chr19_+_10654261 | 0.11 |
ENST00000449870.5
|
ILF3
|
interleukin enhancer binding factor 3 |
| chr7_+_76424922 | 0.11 |
ENST00000394857.8
|
ZP3
|
zona pellucida glycoprotein 3 |
| chr4_-_86101922 | 0.11 |
ENST00000472236.5
ENST00000641881.1 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr11_-_102798148 | 0.11 |
ENST00000315274.7
|
MMP1
|
matrix metallopeptidase 1 |
| chr1_+_13060769 | 0.11 |
ENST00000617807.3
|
HNRNPCL3
|
heterogeneous nuclear ribonucleoprotein C like 3 |
| chr7_+_139829242 | 0.11 |
ENST00000455353.6
ENST00000458722.6 ENST00000448866.7 ENST00000411653.6 |
TBXAS1
|
thromboxane A synthase 1 |
| chr1_-_205775182 | 0.11 |
ENST00000446390.6
|
RAB29
|
RAB29, member RAS oncogene family |
| chr6_-_42142604 | 0.11 |
ENST00000356542.5
ENST00000341865.9 |
C6orf132
|
chromosome 6 open reading frame 132 |
| chrY_+_2841864 | 0.11 |
ENST00000430575.1
|
RPS4Y1
|
ribosomal protein S4 Y-linked 1 |
| chr12_-_8662881 | 0.11 |
ENST00000433590.6
|
MFAP5
|
microfibril associated protein 5 |
| chr9_+_132978687 | 0.11 |
ENST00000372122.4
ENST00000372123.5 |
GFI1B
|
growth factor independent 1B transcriptional repressor |
| chr3_+_178419123 | 0.11 |
ENST00000614557.1
ENST00000455307.5 ENST00000436432.1 |
ENSG00000275163.1
LINC01014
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 (KCNMB2-IT1 - KCNMB2 readthrough transcript) long intergenic non-protein coding RNA 1014 |
| chr16_+_10386049 | 0.11 |
ENST00000562527.5
ENST00000396559.5 ENST00000396560.6 ENST00000562102.5 ENST00000543967.5 ENST00000569939.5 ENST00000569900.5 |
ATF7IP2
|
activating transcription factor 7 interacting protein 2 |
| chr2_+_233729042 | 0.11 |
ENST00000482026.6
|
UGT1A3
|
UDP glucuronosyltransferase family 1 member A3 |
| chr19_+_10654327 | 0.10 |
ENST00000407004.7
ENST00000589998.5 ENST00000589600.5 |
ILF3
|
interleukin enhancer binding factor 3 |
| chr7_+_135557904 | 0.10 |
ENST00000285968.11
|
NUP205
|
nucleoporin 205 |
| chr15_-_42920638 | 0.10 |
ENST00000566931.1
ENST00000564431.5 ENST00000567274.5 ENST00000267890.11 |
TTBK2
|
tau tubulin kinase 2 |
| chr14_+_50872098 | 0.10 |
ENST00000353130.5
|
ABHD12B
|
abhydrolase domain containing 12B |
| chr9_+_69123009 | 0.10 |
ENST00000647986.1
|
TJP2
|
tight junction protein 2 |
| chr11_+_60378524 | 0.10 |
ENST00000530614.5
ENST00000530027.5 ENST00000300184.8 ENST00000530234.2 ENST00000528215.1 ENST00000531787.5 |
MS4A7
MS4A14
|
membrane spanning 4-domains A7 membrane spanning 4-domains A14 |
| chr15_-_33067884 | 0.10 |
ENST00000334528.13
|
FMN1
|
formin 1 |
| chr6_-_32953017 | 0.10 |
ENST00000395305.7
ENST00000374843.9 ENST00000395303.7 ENST00000429234.1 |
HLA-DMA
ENSG00000248993.1
|
major histocompatibility complex, class II, DM alpha novel protein |
| chr5_+_44808930 | 0.10 |
ENST00000507110.6
|
MRPS30
|
mitochondrial ribosomal protein S30 |
| chr5_-_175961324 | 0.10 |
ENST00000432305.6
ENST00000505969.1 |
THOC3
|
THO complex 3 |
| chr18_+_3451585 | 0.10 |
ENST00000551541.5
|
TGIF1
|
TGFB induced factor homeobox 1 |
| chr7_+_142670734 | 0.10 |
ENST00000390398.3
|
TRBV25-1
|
T cell receptor beta variable 25-1 |
| chr1_+_173824694 | 0.09 |
ENST00000647645.1
|
DARS2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr11_-_118225002 | 0.09 |
ENST00000356289.10
ENST00000526620.5 |
JAML
|
junction adhesion molecule like |
| chr17_-_75855204 | 0.09 |
ENST00000589642.5
ENST00000593002.1 ENST00000590221.5 ENST00000587374.5 ENST00000585462.5 ENST00000254806.8 ENST00000433525.6 ENST00000626827.2 |
WBP2
|
WW domain binding protein 2 |
| chr14_-_54902807 | 0.09 |
ENST00000543643.6
ENST00000536224.2 ENST00000395514.5 ENST00000491895.7 |
GCH1
|
GTP cyclohydrolase 1 |
| chr19_-_49453472 | 0.09 |
ENST00000601825.1
ENST00000596049.5 ENST00000599366.5 ENST00000597415.5 |
PIH1D1
|
PIH1 domain containing 1 |
| chr15_-_42920798 | 0.09 |
ENST00000622375.4
ENST00000567840.5 |
TTBK2
|
tau tubulin kinase 2 |
| chr1_+_173824626 | 0.09 |
ENST00000648960.1
ENST00000648807.1 ENST00000649067.1 ENST00000649689.2 |
DARS2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr20_+_1203454 | 0.09 |
ENST00000400633.2
|
C20orf202
|
chromosome 20 open reading frame 202 |
| chrX_-_104013708 | 0.09 |
ENST00000217926.7
|
H2BW1
|
H2B.W histone 1 |
| chr18_+_9885726 | 0.09 |
ENST00000611534.1
ENST00000536353.2 ENST00000584255.1 |
TXNDC2
|
thioredoxin domain containing 2 |
| chr1_+_13061158 | 0.09 |
ENST00000681473.1
|
HNRNPCL3
|
heterogeneous nuclear ribonucleoprotein C like 3 |
| chr1_-_13165631 | 0.08 |
ENST00000323770.8
|
HNRNPCL4
|
heterogeneous nuclear ribonucleoprotein C like 4 |
| chr16_-_30012294 | 0.08 |
ENST00000564979.5
ENST00000563378.5 |
DOC2A
|
double C2 domain alpha |
| chrX_+_8464830 | 0.08 |
ENST00000453306.4
ENST00000381032.6 ENST00000444481.3 |
VCX3B
|
variable charge X-linked 3B |
| chr5_+_139342442 | 0.08 |
ENST00000394795.6
ENST00000510080.1 |
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr22_+_32357867 | 0.08 |
ENST00000249007.4
|
RFPL3
|
ret finger protein like 3 |
| chr16_-_20697680 | 0.08 |
ENST00000520010.6
|
ACSM1
|
acyl-CoA synthetase medium chain family member 1 |
| chr3_-_48440022 | 0.08 |
ENST00000447018.5
ENST00000442740.1 ENST00000412398.6 ENST00000395694.7 |
CCDC51
|
coiled-coil domain containing 51 |
| chr2_+_233617626 | 0.08 |
ENST00000373450.5
|
UGT1A8
|
UDP glucuronosyltransferase family 1 member A8 |
| chr2_+_1414382 | 0.08 |
ENST00000423320.5
ENST00000346956.7 ENST00000382198.5 |
TPO
|
thyroid peroxidase |
| chr1_+_53894181 | 0.08 |
ENST00000361921.8
ENST00000322679.10 ENST00000613679.4 ENST00000617230.2 ENST00000610607.4 ENST00000532493.5 ENST00000525202.5 ENST00000524406.5 ENST00000388876.3 |
DIO1
|
iodothyronine deiodinase 1 |
| chr17_-_41489907 | 0.08 |
ENST00000328119.11
|
KRT36
|
keratin 36 |
| chr6_-_143450662 | 0.08 |
ENST00000237283.9
|
ADAT2
|
adenosine deaminase tRNA specific 2 |
| chr10_-_125683155 | 0.08 |
ENST00000368821.4
|
TEX36
|
testis expressed 36 |
| chr18_+_23689439 | 0.08 |
ENST00000313654.14
|
LAMA3
|
laminin subunit alpha 3 |
| chr18_+_63702958 | 0.08 |
ENST00000544088.6
|
SERPINB11
|
serpin family B member 11 |
| chr1_+_151036578 | 0.08 |
ENST00000368931.8
ENST00000295294.11 |
BNIPL
|
BCL2 interacting protein like |
| chr3_-_193378747 | 0.08 |
ENST00000342358.9
|
ATP13A5
|
ATPase 13A5 |
| chr3_+_185586270 | 0.08 |
ENST00000296257.10
|
SENP2
|
SUMO specific peptidase 2 |
| chr16_-_15094008 | 0.07 |
ENST00000327307.11
|
RRN3
|
RRN3 homolog, RNA polymerase I transcription factor |
| chr14_-_106511856 | 0.07 |
ENST00000390622.2
|
IGHV1-46
|
immunoglobulin heavy variable 1-46 |
| chr11_+_60378494 | 0.07 |
ENST00000534016.5
|
MS4A7
|
membrane spanning 4-domains A7 |
| chr11_-_7674206 | 0.07 |
ENST00000533558.5
ENST00000527542.5 |
CYB5R2
|
cytochrome b5 reductase 2 |
| chr2_-_156342348 | 0.07 |
ENST00000409572.5
|
NR4A2
|
nuclear receptor subfamily 4 group A member 2 |
| chr3_+_98732236 | 0.07 |
ENST00000265261.10
ENST00000483910.5 ENST00000460774.5 |
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr9_-_21385395 | 0.07 |
ENST00000380206.4
|
IFNA2
|
interferon alpha 2 |
| chr6_+_7590179 | 0.07 |
ENST00000342415.6
|
SNRNP48
|
small nuclear ribonucleoprotein U11/U12 subunit 48 |
| chr17_-_75289212 | 0.07 |
ENST00000582778.1
ENST00000581988.5 ENST00000579207.5 ENST00000583332.5 ENST00000442286.6 ENST00000580151.5 ENST00000580994.5 ENST00000584438.1 ENST00000416858.7 ENST00000320362.7 |
SLC25A19
|
solute carrier family 25 member 19 |
| chr11_+_5351508 | 0.07 |
ENST00000380219.1
|
OR51B6
|
olfactory receptor family 51 subfamily B member 6 |
| chrY_-_23694579 | 0.07 |
ENST00000343584.10
|
PRYP3
|
PTPN13 like Y-linked pseudogene 3 |
| chr15_-_89211803 | 0.07 |
ENST00000563254.1
|
RLBP1
|
retinaldehyde binding protein 1 |
| chr19_+_16661121 | 0.07 |
ENST00000187762.7
ENST00000599479.1 |
TMEM38A
|
transmembrane protein 38A |
| chrX_-_103064164 | 0.07 |
ENST00000372728.4
|
BEX1
|
brain expressed X-linked 1 |
| chr1_-_205121986 | 0.07 |
ENST00000367164.1
|
RBBP5
|
RB binding protein 5, histone lysine methyltransferase complex subunit |
| chr6_-_55875583 | 0.07 |
ENST00000370830.4
|
BMP5
|
bone morphogenetic protein 5 |
| chr12_-_7503841 | 0.06 |
ENST00000359156.8
|
CD163
|
CD163 molecule |
| chr6_+_125956696 | 0.06 |
ENST00000229633.7
|
HINT3
|
histidine triad nucleotide binding protein 3 |
| chr1_+_40374648 | 0.06 |
ENST00000372708.5
|
SMAP2
|
small ArfGAP2 |
| chr9_-_122913299 | 0.06 |
ENST00000373659.4
|
ZBTB6
|
zinc finger and BTB domain containing 6 |
| chr6_+_83859640 | 0.06 |
ENST00000369679.4
ENST00000369681.10 |
CYB5R4
|
cytochrome b5 reductase 4 |
| chr3_+_98732688 | 0.06 |
ENST00000486334.6
ENST00000394162.5 ENST00000613264.4 |
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr20_-_63568074 | 0.06 |
ENST00000427522.6
|
HELZ2
|
helicase with zinc finger 2 |
| chr7_+_95485898 | 0.06 |
ENST00000428113.5
|
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr14_+_103563171 | 0.06 |
ENST00000492189.5
ENST00000477116.5 ENST00000473127.5 |
COA8
|
cytochrome c oxidase assembly factor 8 |
| chr14_+_20768393 | 0.06 |
ENST00000326783.4
|
EDDM3B
|
epididymal protein 3B |
| chr2_-_88947820 | 0.06 |
ENST00000496168.1
|
IGKV1-5
|
immunoglobulin kappa variable 1-5 |
| chr1_+_54209946 | 0.06 |
ENST00000398219.2
|
MRPL37
|
mitochondrial ribosomal protein L37 |
| chr19_-_15479469 | 0.06 |
ENST00000292609.8
ENST00000340880.5 |
PGLYRP2
|
peptidoglycan recognition protein 2 |
| chr8_+_22995831 | 0.06 |
ENST00000522948.5
|
RHOBTB2
|
Rho related BTB domain containing 2 |
| chr1_+_205256189 | 0.06 |
ENST00000329800.7
|
TMCC2
|
transmembrane and coiled-coil domain family 2 |
| chr11_+_108008880 | 0.06 |
ENST00000393094.7
|
CUL5
|
cullin 5 |
| chr20_-_10420737 | 0.06 |
ENST00000649912.1
|
ENSG00000285723.1
|
novel protein |
| chr1_+_86468902 | 0.06 |
ENST00000394711.2
|
CLCA1
|
chloride channel accessory 1 |
| chr5_+_139341875 | 0.06 |
ENST00000511706.5
|
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr14_+_103563270 | 0.06 |
ENST00000476323.5
|
COA8
|
cytochrome c oxidase assembly factor 8 |
| chr14_-_105065422 | 0.05 |
ENST00000329797.8
ENST00000539291.6 ENST00000392585.2 |
GPR132
|
G protein-coupled receptor 132 |
| chr6_-_49787265 | 0.05 |
ENST00000304801.6
|
PGK2
|
phosphoglycerate kinase 2 |
| chr22_+_24555981 | 0.05 |
ENST00000215829.8
ENST00000402849.5 |
SNRPD3
|
small nuclear ribonucleoprotein D3 polypeptide |
| chr11_-_57649932 | 0.05 |
ENST00000524669.5
|
YPEL4
|
yippee like 4 |
| chr17_+_58166982 | 0.05 |
ENST00000545221.2
|
OR4D2
|
olfactory receptor family 4 subfamily D member 2 |
| chr2_+_233671879 | 0.05 |
ENST00000354728.5
|
UGT1A9
|
UDP glucuronosyltransferase family 1 member A9 |
| chr11_-_35418966 | 0.05 |
ENST00000531628.2
|
SLC1A2
|
solute carrier family 1 member 2 |
| chr15_-_76012390 | 0.05 |
ENST00000394907.8
|
NRG4
|
neuregulin 4 |
| chr17_+_27471999 | 0.05 |
ENST00000583370.5
ENST00000509603.6 ENST00000268763.10 ENST00000398988.7 |
KSR1
|
kinase suppressor of ras 1 |
| chr1_-_205121964 | 0.05 |
ENST00000264515.11
|
RBBP5
|
RB binding protein 5, histone lysine methyltransferase complex subunit |
| chr12_-_7502730 | 0.05 |
ENST00000541972.5
|
CD163
|
CD163 molecule |
| chr14_-_106557465 | 0.05 |
ENST00000390625.3
|
IGHV3-49
|
immunoglobulin heavy variable 3-49 |
| chr5_+_115841878 | 0.05 |
ENST00000316788.12
|
AP3S1
|
adaptor related protein complex 3 subunit sigma 1 |
| chr8_-_140800535 | 0.05 |
ENST00000521986.5
ENST00000523539.5 |
PTK2
|
protein tyrosine kinase 2 |
| chrX_+_49331616 | 0.05 |
ENST00000612958.1
|
GAGE13
|
G antigen 13 |
| chr1_+_197413827 | 0.05 |
ENST00000367397.1
ENST00000681519.1 |
CRB1
|
crumbs cell polarity complex component 1 |
| chr5_-_16508990 | 0.05 |
ENST00000399793.6
|
RETREG1
|
reticulophagy regulator 1 |
| chr18_+_22933819 | 0.05 |
ENST00000399722.6
ENST00000399725.6 ENST00000399721.6 |
RBBP8
|
RB binding protein 8, endonuclease |
| chr1_+_108877135 | 0.05 |
ENST00000675086.1
ENST00000676184.1 ENST00000675087.1 |
GPSM2
|
G protein signaling modulator 2 |
| chr6_-_111605859 | 0.05 |
ENST00000651359.1
ENST00000650859.1 ENST00000359831.8 ENST00000368761.11 |
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr19_-_3626746 | 0.05 |
ENST00000429344.7
ENST00000248420.9 ENST00000221899.7 |
CACTIN
|
cactin, spliceosome C complex subunit |
| chr6_-_8102481 | 0.05 |
ENST00000502429.5
ENST00000429723.6 ENST00000379715.10 ENST00000507463.1 |
EEF1E1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr5_-_10307821 | 0.05 |
ENST00000296658.4
|
CMBL
|
carboxymethylenebutenolidase homolog |
| chrX_+_49341192 | 0.05 |
ENST00000621907.1
|
GAGE2E
|
G antigen 2E |
| chr12_-_7503744 | 0.05 |
ENST00000396620.7
ENST00000432237.3 |
CD163
|
CD163 molecule |
| chr6_+_26216928 | 0.05 |
ENST00000303910.4
|
H2AC8
|
H2A clustered histone 8 |
| chr10_-_49269 | 0.05 |
ENST00000562809.1
ENST00000568866.5 ENST00000561967.1 ENST00000568584.6 |
TUBB8
|
tubulin beta 8 class VIII |
| chr7_+_141790217 | 0.05 |
ENST00000247883.5
|
TAS2R5
|
taste 2 receptor member 5 |
| chr3_+_125969152 | 0.05 |
ENST00000251776.8
ENST00000504401.1 |
ROPN1B
|
rhophilin associated tail protein 1B |
| chr16_-_81096336 | 0.05 |
ENST00000639169.1
ENST00000315467.9 |
GCSH
|
glycine cleavage system protein H |
| chr10_+_70052582 | 0.05 |
ENST00000676699.1
|
MACROH2A2
|
macroH2A.2 histone |
| chrX_+_49322057 | 0.05 |
ENST00000442437.2
|
GAGE12J
|
G antigen 12J |
| chr4_+_177309866 | 0.04 |
ENST00000264596.4
|
NEIL3
|
nei like DNA glycosylase 3 |
| chr3_+_58008350 | 0.04 |
ENST00000490882.5
ENST00000358537.7 ENST00000429972.6 ENST00000682871.1 ENST00000295956.9 |
FLNB
|
filamin B |
| chr7_-_6348906 | 0.04 |
ENST00000313324.9
ENST00000530143.1 |
FAM220A
|
family with sequence similarity 220 member A |
| chr6_-_125301900 | 0.04 |
ENST00000608295.5
ENST00000398153.7 ENST00000608284.1 |
HDDC2
|
HD domain containing 2 |
| chr16_+_33827140 | 0.04 |
ENST00000562905.2
|
IGHV3OR16-13
|
immunoglobulin heavy variable 3/OR16-13 (non-functional) |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX17
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 | 0.4 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 0.4 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.1 | 0.2 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.1 | 0.3 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.1 | 0.4 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.1 | 0.2 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 0.3 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.3 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.0 | 0.4 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 2.0 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.2 | GO:0050904 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) diapedesis(GO:0050904) |
| 0.0 | 0.1 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.1 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.0 | 0.4 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.1 | GO:1900110 | negative regulation of histone H3-K9 dimethylation(GO:1900110) regulation of TORC2 signaling(GO:1903939) |
| 0.0 | 0.1 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.0 | 0.1 | GO:2000360 | positive regulation of female gonad development(GO:2000196) negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.0 | 0.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.3 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.1 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.0 | 0.2 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:0030974 | thiamine pyrophosphate transport(GO:0030974) |
| 0.0 | 0.1 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.0 | 0.2 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.1 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.2 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.1 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.1 | GO:0032827 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.0 | 0.2 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.2 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.0 | 0.1 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.2 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.1 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.0 | 0.1 | GO:0035696 | monocyte extravasation(GO:0035696) |
| 0.0 | 0.1 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.1 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.0 | 0.2 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.0 | GO:1904692 | positive regulation of type B pancreatic cell proliferation(GO:1904692) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.1 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) regulation of lung blood pressure(GO:0014916) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.3 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 0.1 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 0.1 | GO:2000662 | interleukin-5 secretion(GO:0072603) regulation of interleukin-5 secretion(GO:2000662) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.4 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.2 | GO:0005713 | recombination nodule(GO:0005713) |
| 0.0 | 0.2 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 0.2 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0020003 | symbiont-containing vacuole(GO:0020003) |
| 0.0 | 0.4 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.1 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.0 | 0.4 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.0 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.0 | 0.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.1 | 0.4 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.1 | 0.4 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.1 | 0.3 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.1 | 0.2 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 0.3 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.1 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.1 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.0 | 0.3 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.2 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.1 | GO:0090422 | thiamine pyrophosphate transporter activity(GO:0090422) |
| 0.0 | 0.1 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.0 | 0.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.2 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.1 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.4 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.0 | 0.1 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 2.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.0 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.1 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 0.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.2 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) acrosin binding(GO:0032190) |
| 0.0 | 0.0 | GO:0097677 | STAT family protein binding(GO:0097677) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.0 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.5 | PID ATM PATHWAY | ATM pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.6 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |