Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
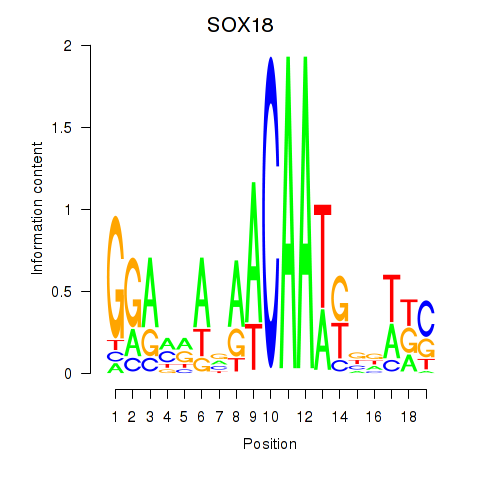
Results for SOX18
Z-value: 0.51
Transcription factors associated with SOX18
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX18
|
ENSG00000203883.7 | SRY-box transcription factor 18 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX18 | hg38_v1_chr20_-_64049631_64049646 | -0.22 | 6.0e-01 | Click! |
Activity profile of SOX18 motif
Sorted Z-values of SOX18 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_101452629 | 0.49 |
ENST00000622834.4
ENST00000545560.6 ENST00000376180.8 |
ITGBL1
|
integrin subunit beta like 1 |
| chr11_+_5689780 | 0.41 |
ENST00000379965.8
ENST00000454828.5 |
TRIM22
|
tripartite motif containing 22 |
| chr2_+_32946944 | 0.36 |
ENST00000404816.7
|
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr1_-_72100930 | 0.34 |
ENST00000306821.3
|
NEGR1
|
neuronal growth regulator 1 |
| chr2_-_37672178 | 0.31 |
ENST00000457889.1
|
CDC42EP3
|
CDC42 effector protein 3 |
| chr17_+_43006740 | 0.31 |
ENST00000438323.2
ENST00000415816.7 |
IFI35
|
interferon induced protein 35 |
| chr7_+_142111739 | 0.30 |
ENST00000550469.6
ENST00000477922.3 |
MGAM2
|
maltase-glucoamylase 2 (putative) |
| chr21_-_30372265 | 0.28 |
ENST00000399889.4
|
KRTAP13-2
|
keratin associated protein 13-2 |
| chr8_-_13276491 | 0.27 |
ENST00000512044.6
|
DLC1
|
DLC1 Rho GTPase activating protein |
| chr9_-_121050264 | 0.26 |
ENST00000223642.3
|
C5
|
complement C5 |
| chr13_-_33205997 | 0.26 |
ENST00000399365.7
|
STARD13
|
StAR related lipid transfer domain containing 13 |
| chr11_-_89490715 | 0.25 |
ENST00000528341.5
|
NOX4
|
NADPH oxidase 4 |
| chr3_-_112641292 | 0.22 |
ENST00000439685.6
|
CCDC80
|
coiled-coil domain containing 80 |
| chr5_-_137499293 | 0.22 |
ENST00000510689.5
ENST00000394945.6 |
SPOCK1
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 1 |
| chr5_-_76623391 | 0.21 |
ENST00000296641.5
ENST00000504899.1 |
F2RL2
|
coagulation factor II thrombin receptor like 2 |
| chr6_-_152168291 | 0.21 |
ENST00000354674.5
|
SYNE1
|
spectrin repeat containing nuclear envelope protein 1 |
| chr6_-_152168349 | 0.21 |
ENST00000539504.5
|
SYNE1
|
spectrin repeat containing nuclear envelope protein 1 |
| chr1_-_153544997 | 0.19 |
ENST00000368715.5
|
S100A4
|
S100 calcium binding protein A4 |
| chr1_-_94927079 | 0.19 |
ENST00000370206.9
ENST00000394202.8 |
CNN3
|
calponin 3 |
| chr12_-_10435940 | 0.17 |
ENST00000381901.5
ENST00000381902.7 ENST00000539033.1 |
KLRC2
ENSG00000255641.1
|
killer cell lectin like receptor C2 novel protein |
| chr1_-_182391363 | 0.16 |
ENST00000417584.6
|
GLUL
|
glutamate-ammonia ligase |
| chr3_-_121660892 | 0.16 |
ENST00000428394.6
ENST00000314583.8 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr22_-_38794111 | 0.16 |
ENST00000406622.5
ENST00000216068.9 ENST00000406199.3 |
SUN2
DNAL4
|
Sad1 and UNC84 domain containing 2 dynein axonemal light chain 4 |
| chr14_-_106811131 | 0.15 |
ENST00000424969.2
|
IGHV3-74
|
immunoglobulin heavy variable 3-74 |
| chr2_-_37671633 | 0.15 |
ENST00000295324.4
|
CDC42EP3
|
CDC42 effector protein 3 |
| chr11_-_134253248 | 0.15 |
ENST00000392595.6
ENST00000352327.5 ENST00000341541.8 ENST00000392594.7 |
THYN1
|
thymocyte nuclear protein 1 |
| chr1_-_248645278 | 0.15 |
ENST00000641268.1
|
OR2T35
|
olfactory receptor family 2 subfamily T member 35 |
| chr1_-_182391783 | 0.15 |
ENST00000331872.11
ENST00000339526.8 |
GLUL
|
glutamate-ammonia ligase |
| chr4_+_108620567 | 0.15 |
ENST00000502534.5
ENST00000394667.8 |
RPL34
|
ribosomal protein L34 |
| chr20_-_46089905 | 0.14 |
ENST00000372291.3
ENST00000290231.11 |
NCOA5
|
nuclear receptor coactivator 5 |
| chr10_-_125816596 | 0.14 |
ENST00000368786.5
|
UROS
|
uroporphyrinogen III synthase |
| chr7_-_93148345 | 0.14 |
ENST00000437805.5
ENST00000446959.5 ENST00000439952.5 ENST00000414791.5 ENST00000446033.1 ENST00000411955.5 ENST00000318238.9 |
SAMD9L
|
sterile alpha motif domain containing 9 like |
| chr12_+_15322529 | 0.13 |
ENST00000348962.7
|
PTPRO
|
protein tyrosine phosphatase receptor type O |
| chr12_-_10420550 | 0.12 |
ENST00000381903.2
ENST00000396439.7 |
KLRC3
|
killer cell lectin like receptor C3 |
| chr3_-_93973833 | 0.12 |
ENST00000348974.5
ENST00000647936.1 ENST00000394236.9 ENST00000407433.6 |
PROS1
|
protein S |
| chr11_-_89491131 | 0.12 |
ENST00000343727.9
ENST00000531342.5 ENST00000375979.7 |
NOX4
|
NADPH oxidase 4 |
| chr22_-_44498179 | 0.11 |
ENST00000341255.4
|
RTL6
|
retrotransposon Gag like 6 |
| chr3_-_93973933 | 0.11 |
ENST00000650591.1
|
PROS1
|
protein S |
| chr5_+_141392616 | 0.11 |
ENST00000398604.3
|
PCDHGA8
|
protocadherin gamma subfamily A, 8 |
| chr1_+_201780490 | 0.11 |
ENST00000430015.5
|
NAV1
|
neuron navigator 1 |
| chr11_-_7941708 | 0.10 |
ENST00000642047.1
|
OR10A3
|
olfactory receptor family 10 subfamily A member 3 |
| chr5_+_141343818 | 0.10 |
ENST00000619750.1
ENST00000253812.8 |
PCDHGA3
|
protocadherin gamma subfamily A, 3 |
| chr15_-_64989435 | 0.10 |
ENST00000433215.6
ENST00000558415.5 ENST00000557795.5 |
SPG21
|
SPG21 abhydrolase domain containing, maspardin |
| chr22_+_42553862 | 0.09 |
ENST00000340239.8
ENST00000327678.10 |
SERHL2
|
serine hydrolase like 2 |
| chr6_-_26234978 | 0.09 |
ENST00000244534.7
|
H1-3
|
H1.3 linker histone, cluster member |
| chr4_+_108620616 | 0.09 |
ENST00000506397.5
ENST00000394668.2 |
RPL34
|
ribosomal protein L34 |
| chr4_+_108620584 | 0.09 |
ENST00000394665.5
|
RPL34
|
ribosomal protein L34 |
| chr2_-_210303608 | 0.09 |
ENST00000341685.8
|
MYL1
|
myosin light chain 1 |
| chr1_-_11858935 | 0.09 |
ENST00000376468.4
|
NPPB
|
natriuretic peptide B |
| chr15_+_43791842 | 0.09 |
ENST00000674451.1
ENST00000630046.2 |
SERF2
|
small EDRK-rich factor 2 |
| chr22_-_38084093 | 0.08 |
ENST00000681075.1
|
SLC16A8
|
solute carrier family 16 member 8 |
| chr16_-_4474522 | 0.08 |
ENST00000573571.1
|
NMRAL1
|
NmrA like redox sensor 1 |
| chr16_-_4846196 | 0.08 |
ENST00000589389.5
|
GLYR1
|
glyoxylate reductase 1 homolog |
| chr9_-_96302104 | 0.08 |
ENST00000375262.4
ENST00000650386.1 |
HSD17B3
|
hydroxysteroid 17-beta dehydrogenase 3 |
| chr7_-_6009072 | 0.08 |
ENST00000642456.1
ENST00000642292.1 ENST00000441476.6 |
PMS2
|
PMS1 homolog 2, mismatch repair system component |
| chr12_-_110502065 | 0.08 |
ENST00000447578.6
ENST00000546588.1 ENST00000360579.11 ENST00000549578.6 ENST00000549970.5 |
VPS29
|
VPS29 retromer complex component |
| chr18_-_5396265 | 0.08 |
ENST00000579951.2
|
EPB41L3
|
erythrocyte membrane protein band 4.1 like 3 |
| chr22_+_44026283 | 0.08 |
ENST00000619710.4
|
PARVB
|
parvin beta |
| chr14_+_23988914 | 0.08 |
ENST00000335125.11
|
DHRS4L2
|
dehydrogenase/reductase 4 like 2 |
| chr7_+_12687625 | 0.08 |
ENST00000651779.1
ENST00000404894.1 |
ARL4A
|
ADP ribosylation factor like GTPase 4A |
| chr5_-_115262851 | 0.08 |
ENST00000379615.3
ENST00000419445.6 |
PGGT1B
|
protein geranylgeranyltransferase type I subunit beta |
| chr2_+_209653171 | 0.08 |
ENST00000447185.5
|
MAP2
|
microtubule associated protein 2 |
| chr3_+_133746385 | 0.08 |
ENST00000482271.5
ENST00000402696.9 |
TF
|
transferrin |
| chr3_+_156674579 | 0.07 |
ENST00000295924.12
|
TIPARP
|
TCDD inducible poly(ADP-ribose) polymerase |
| chr14_+_23988884 | 0.07 |
ENST00000558753.5
ENST00000537912.5 |
DHRS4L2
|
dehydrogenase/reductase 4 like 2 |
| chr16_+_66603874 | 0.07 |
ENST00000563672.5
ENST00000424011.6 |
CMTM3
|
CKLF like MARVEL transmembrane domain containing 3 |
| chr5_+_141387698 | 0.07 |
ENST00000615384.1
ENST00000519479.2 |
PCDHGB4
|
protocadherin gamma subfamily B, 4 |
| chr16_-_15643024 | 0.07 |
ENST00000540441.6
|
MARF1
|
meiosis regulator and mRNA stability factor 1 |
| chr13_-_52450590 | 0.07 |
ENST00000378060.9
|
VPS36
|
vacuolar protein sorting 36 homolog |
| chrX_-_154805386 | 0.07 |
ENST00000393531.5
ENST00000369534.8 ENST00000453245.5 ENST00000428488.1 ENST00000369531.1 |
MPP1
|
membrane palmitoylated protein 1 |
| chr1_-_156248013 | 0.07 |
ENST00000368270.2
|
PAQR6
|
progestin and adipoQ receptor family member 6 |
| chr1_+_9239835 | 0.07 |
ENST00000602477.1
|
H6PD
|
hexose-6-phosphate dehydrogenase/glucose 1-dehydrogenase |
| chr12_+_2877193 | 0.07 |
ENST00000538636.5
ENST00000461997.5 ENST00000618250.4 ENST00000366285.5 ENST00000538700.2 ENST00000489288.7 |
RHNO1
|
RAD9-HUS1-RAD1 interacting nuclear orphan 1 |
| chr5_+_32711723 | 0.07 |
ENST00000415167.2
|
NPR3
|
natriuretic peptide receptor 3 |
| chr19_+_44072142 | 0.07 |
ENST00000421176.4
|
ZNF284
|
zinc finger protein 284 |
| chr5_+_141421064 | 0.07 |
ENST00000518882.2
|
PCDHGA11
|
protocadherin gamma subfamily A, 11 |
| chr1_+_161153968 | 0.07 |
ENST00000368003.6
|
UFC1
|
ubiquitin-fold modifier conjugating enzyme 1 |
| chr12_+_95858928 | 0.07 |
ENST00000266735.9
ENST00000553192.5 ENST00000552085.1 |
SNRPF
|
small nuclear ribonucleoprotein polypeptide F |
| chr1_-_155911365 | 0.07 |
ENST00000651833.1
ENST00000539040.5 ENST00000651853.1 |
RIT1
|
Ras like without CAAX 1 |
| chr12_-_55927865 | 0.07 |
ENST00000454792.2
ENST00000408946.7 |
PYM1
|
PYM homolog 1, exon junction complex associated factor |
| chr1_-_155911340 | 0.06 |
ENST00000368323.8
|
RIT1
|
Ras like without CAAX 1 |
| chr19_+_35641728 | 0.06 |
ENST00000619399.4
ENST00000379026.6 ENST00000379023.8 ENST00000402764.6 ENST00000479824.5 |
ETV2
|
ETS variant transcription factor 2 |
| chr12_+_104303726 | 0.06 |
ENST00000527879.2
|
EID3
|
EP300 interacting inhibitor of differentiation 3 |
| chr7_-_135977308 | 0.06 |
ENST00000435723.1
ENST00000393085.4 |
MTPN
|
myotrophin |
| chr12_-_68253502 | 0.06 |
ENST00000328087.6
ENST00000538666.6 |
IL22
|
interleukin 22 |
| chr10_-_13348270 | 0.06 |
ENST00000378614.8
ENST00000327347.10 ENST00000545675.5 |
SEPHS1
|
selenophosphate synthetase 1 |
| chr18_+_3447562 | 0.06 |
ENST00000618001.4
|
TGIF1
|
TGFB induced factor homeobox 1 |
| chr1_-_156248084 | 0.06 |
ENST00000652405.1
ENST00000335852.6 ENST00000540423.5 ENST00000612424.4 ENST00000613336.4 ENST00000623241.3 |
PAQR6
|
progestin and adipoQ receptor family member 6 |
| chr4_+_153152163 | 0.06 |
ENST00000676423.1
ENST00000675745.1 ENST00000676348.1 ENST00000676408.1 ENST00000674874.1 ENST00000675315.1 ENST00000675518.1 |
TRIM2
ENSG00000288637.1
|
tripartite motif containing 2 novel protein |
| chr8_+_23288081 | 0.06 |
ENST00000265806.12
ENST00000519952.6 ENST00000518840.6 |
R3HCC1
|
R3H domain and coiled-coil containing 1 |
| chr1_-_156248038 | 0.06 |
ENST00000470198.5
ENST00000292291.10 ENST00000356983.7 |
PAQR6
|
progestin and adipoQ receptor family member 6 |
| chr2_+_44275473 | 0.06 |
ENST00000260649.11
|
SLC3A1
|
solute carrier family 3 member 1 |
| chr14_-_70416994 | 0.06 |
ENST00000621525.4
ENST00000256366.6 |
SYNJ2BP-COX16
SYNJ2BP
|
SYNJ2BP-COX16 readthrough synaptojanin 2 binding protein |
| chr3_-_146460440 | 0.06 |
ENST00000610787.5
|
PLSCR2
|
phospholipid scramblase 2 |
| chr4_-_7042931 | 0.06 |
ENST00000310085.6
|
CCDC96
|
coiled-coil domain containing 96 |
| chr5_+_31532277 | 0.06 |
ENST00000507818.6
ENST00000325366.14 |
C5orf22
|
chromosome 5 open reading frame 22 |
| chr3_+_19148500 | 0.06 |
ENST00000328405.7
|
KCNH8
|
potassium voltage-gated channel subfamily H member 8 |
| chr19_+_57231014 | 0.06 |
ENST00000302804.12
|
AURKC
|
aurora kinase C |
| chr1_+_207089283 | 0.06 |
ENST00000391923.1
|
C4BPB
|
complement component 4 binding protein beta |
| chrX_+_47218232 | 0.06 |
ENST00000457458.6
ENST00000522883.1 |
CDK16
|
cyclin dependent kinase 16 |
| chr19_+_57231001 | 0.06 |
ENST00000415300.6
|
AURKC
|
aurora kinase C |
| chrX_+_86714623 | 0.06 |
ENST00000484479.1
|
DACH2
|
dachshund family transcription factor 2 |
| chr2_+_105337515 | 0.06 |
ENST00000410049.1
ENST00000258457.7 |
C2orf49
|
chromosome 2 open reading frame 49 |
| chr11_+_60040402 | 0.05 |
ENST00000278855.7
ENST00000532905.1 |
OOSP2
|
oocyte secreted protein 2 |
| chr16_-_1611985 | 0.05 |
ENST00000426508.7
|
IFT140
|
intraflagellar transport 140 |
| chr15_+_62066975 | 0.05 |
ENST00000355522.5
|
C2CD4A
|
C2 calcium dependent domain containing 4A |
| chr22_+_37282464 | 0.05 |
ENST00000402997.5
ENST00000405206.3 ENST00000248901.11 |
CYTH4
|
cytohesin 4 |
| chr1_-_155910881 | 0.05 |
ENST00000609492.1
ENST00000368322.7 |
RIT1
|
Ras like without CAAX 1 |
| chr2_-_61017174 | 0.05 |
ENST00000407787.5
ENST00000398658.2 |
PUS10
|
pseudouridine synthase 10 |
| chr7_-_93574721 | 0.05 |
ENST00000426151.7
ENST00000649521.1 |
CALCR
|
calcitonin receptor |
| chrX_+_47218670 | 0.05 |
ENST00000357227.9
ENST00000519758.5 ENST00000520893.5 ENST00000622098.4 ENST00000517426.5 |
CDK16
|
cyclin dependent kinase 16 |
| chr17_+_28371656 | 0.05 |
ENST00000585482.6
|
SARM1
|
sterile alpha and TIR motif containing 1 |
| chr1_-_165445220 | 0.05 |
ENST00000619224.1
|
RXRG
|
retinoid X receptor gamma |
| chr7_+_134866831 | 0.05 |
ENST00000435928.1
|
CALD1
|
caldesmon 1 |
| chr19_-_35745391 | 0.05 |
ENST00000378975.8
ENST00000587886.2 ENST00000412391.6 ENST00000292879.9 |
U2AF1L4
|
U2 small nuclear RNA auxiliary factor 1 like 4 |
| chr19_+_54630410 | 0.05 |
ENST00000396327.7
ENST00000324602.12 |
LILRB1
|
leukocyte immunoglobulin like receptor B1 |
| chr6_-_111793871 | 0.05 |
ENST00000368667.6
|
FYN
|
FYN proto-oncogene, Src family tyrosine kinase |
| chr1_-_13116854 | 0.05 |
ENST00000621994.3
|
HNRNPCL2
|
heterogeneous nuclear ribonucleoprotein C like 2 |
| chr2_+_241734602 | 0.05 |
ENST00000403782.5
ENST00000321264.9 |
D2HGDH
|
D-2-hydroxyglutarate dehydrogenase |
| chr16_-_4474549 | 0.05 |
ENST00000404295.7
ENST00000283429.11 ENST00000574425.5 |
NMRAL1
|
NmrA like redox sensor 1 |
| chrX_+_101390824 | 0.05 |
ENST00000427805.6
ENST00000614077.4 |
RPL36A
|
ribosomal protein L36a |
| chr6_-_31806937 | 0.05 |
ENST00000375661.6
|
LSM2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr12_-_14938508 | 0.05 |
ENST00000266397.7
|
ERP27
|
endoplasmic reticulum protein 27 |
| chr11_+_196761 | 0.05 |
ENST00000325113.9
|
ODF3
|
outer dense fiber of sperm tails 3 |
| chr5_+_32585549 | 0.04 |
ENST00000265073.9
ENST00000515355.5 ENST00000502897.5 ENST00000510442.1 |
SUB1
|
SUB1 regulator of transcription |
| chr11_+_71527267 | 0.04 |
ENST00000398536.6
|
KRTAP5-7
|
keratin associated protein 5-7 |
| chr15_+_78872809 | 0.04 |
ENST00000331268.9
|
MORF4L1
|
mortality factor 4 like 1 |
| chr12_-_55927756 | 0.04 |
ENST00000549939.1
|
PYM1
|
PYM homolog 1, exon junction complex associated factor |
| chr7_-_6009019 | 0.04 |
ENST00000382321.5
ENST00000265849.12 |
PMS2
|
PMS1 homolog 2, mismatch repair system component |
| chr16_-_29397696 | 0.04 |
ENST00000614927.1
|
NPIPB11
|
nuclear pore complex interacting protein family member B11 |
| chr13_-_51845169 | 0.04 |
ENST00000627246.3
ENST00000629372.3 |
TMEM272
|
transmembrane protein 272 |
| chr10_+_84424919 | 0.04 |
ENST00000543283.2
ENST00000494586.5 |
CCSER2
|
coiled-coil serine rich protein 2 |
| chr4_+_26321365 | 0.04 |
ENST00000505958.6
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr14_-_70416973 | 0.04 |
ENST00000555276.5
ENST00000617124.4 |
COX16
SYNJ2BP-COX16
|
cytochrome c oxidase assembly factor COX16 SYNJ2BP-COX16 readthrough |
| chr1_-_206612436 | 0.04 |
ENST00000437518.1
ENST00000367114.7 ENST00000271764.7 |
EIF2D
|
eukaryotic translation initiation factor 2D |
| chr16_+_22519967 | 0.04 |
ENST00000621622.4
|
NPIPB5
|
nuclear pore complex interacting protein family member B5 |
| chr12_-_110445540 | 0.04 |
ENST00000547365.1
|
ARPC3
|
actin related protein 2/3 complex subunit 3 |
| chr19_-_14835252 | 0.04 |
ENST00000641666.1
ENST00000642030.1 ENST00000642000.1 |
OR7C1
|
olfactory receptor family 7 subfamily C member 1 |
| chr14_+_22070548 | 0.04 |
ENST00000390450.3
|
TRAV22
|
T cell receptor alpha variable 22 |
| chr15_-_74212219 | 0.04 |
ENST00000449139.6
|
STRA6
|
signaling receptor and transporter of retinol STRA6 |
| chr14_+_92794297 | 0.04 |
ENST00000163416.7
|
GOLGA5
|
golgin A5 |
| chr1_+_203861575 | 0.04 |
ENST00000414487.7
|
SNRPE
|
small nuclear ribonucleoprotein polypeptide E |
| chr5_-_67196791 | 0.04 |
ENST00000256447.5
|
CD180
|
CD180 molecule |
| chr11_-_31804067 | 0.04 |
ENST00000639548.1
ENST00000640125.1 ENST00000481563.6 ENST00000639079.1 ENST00000638762.1 ENST00000638346.1 |
PAX6
|
paired box 6 |
| chr1_-_45522870 | 0.04 |
ENST00000424390.2
|
PRDX1
|
peroxiredoxin 1 |
| chr1_-_153608136 | 0.04 |
ENST00000368703.6
|
S100A16
|
S100 calcium binding protein A16 |
| chr15_+_64094060 | 0.04 |
ENST00000560829.5
|
SNX1
|
sorting nexin 1 |
| chr3_+_124384757 | 0.04 |
ENST00000684374.1
|
KALRN
|
kalirin RhoGEF kinase |
| chr12_+_110502307 | 0.03 |
ENST00000409778.7
ENST00000409300.6 |
RAD9B
|
RAD9 checkpoint clamp component B |
| chr4_+_37960397 | 0.03 |
ENST00000504686.2
|
PTTG2
|
pituitary tumor-transforming 2 |
| chr13_-_45418337 | 0.03 |
ENST00000519676.6
ENST00000519547.5 |
SLC25A30
|
solute carrier family 25 member 30 |
| chr12_-_75209422 | 0.03 |
ENST00000393288.2
ENST00000540018.5 |
KCNC2
|
potassium voltage-gated channel subfamily C member 2 |
| chr12_-_110689751 | 0.03 |
ENST00000439744.6
ENST00000549442.5 |
HVCN1
|
hydrogen voltage gated channel 1 |
| chr12_+_110502200 | 0.03 |
ENST00000409246.5
ENST00000392672.8 ENST00000409425.5 |
RAD9B
|
RAD9 checkpoint clamp component B |
| chr11_+_76445001 | 0.03 |
ENST00000533988.5
ENST00000524490.5 ENST00000334736.7 ENST00000533972.5 |
EMSY
|
EMSY transcriptional repressor, BRCA2 interacting |
| chr15_-_74212256 | 0.03 |
ENST00000416286.7
|
STRA6
|
signaling receptor and transporter of retinol STRA6 |
| chr2_+_3658193 | 0.03 |
ENST00000252505.4
|
ALLC
|
allantoicase |
| chr14_-_80231052 | 0.03 |
ENST00000557010.5
|
DIO2
|
iodothyronine deiodinase 2 |
| chr3_-_185923893 | 0.03 |
ENST00000259043.11
|
TRA2B
|
transformer 2 beta homolog |
| chr13_-_44161257 | 0.03 |
ENST00000400419.2
|
SMIM2
|
small integral membrane protein 2 |
| chr15_+_67125707 | 0.03 |
ENST00000540846.6
|
SMAD3
|
SMAD family member 3 |
| chr1_+_42658335 | 0.03 |
ENST00000677900.1
ENST00000372550.6 ENST00000676675.1 ENST00000304979.8 ENST00000678803.1 ENST00000678333.1 ENST00000440068.6 |
PPIH
|
peptidylprolyl isomerase H |
| chr1_+_13303539 | 0.03 |
ENST00000437300.2
|
PRAMEF33
|
PRAME family member 33 |
| chr1_+_64203610 | 0.03 |
ENST00000371077.10
ENST00000611228.4 |
UBE2U
|
ubiquitin conjugating enzyme E2 U |
| chr2_+_79025678 | 0.03 |
ENST00000393897.6
|
REG3G
|
regenerating family member 3 gamma |
| chr11_+_124183219 | 0.03 |
ENST00000641351.2
|
OR10D3
|
olfactory receptor family 10 subfamily D member 3 |
| chr4_+_26321192 | 0.03 |
ENST00000681484.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr4_-_113979635 | 0.03 |
ENST00000315366.8
|
ARSJ
|
arylsulfatase family member J |
| chr14_-_106675544 | 0.03 |
ENST00000390632.2
|
IGHV3-66
|
immunoglobulin heavy variable 3-66 |
| chr4_+_3441960 | 0.03 |
ENST00000382774.8
ENST00000511533.1 |
HGFAC
|
HGF activator |
| chr2_+_233712905 | 0.03 |
ENST00000373414.4
|
UGT1A5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr12_-_663572 | 0.03 |
ENST00000662884.1
|
NINJ2
|
ninjurin 2 |
| chr15_+_21651844 | 0.03 |
ENST00000623441.1
|
OR4N4C
|
olfactory receptor family 4 subfamily N member 4C |
| chr9_-_96302170 | 0.03 |
ENST00000375263.8
|
HSD17B3
|
hydroxysteroid 17-beta dehydrogenase 3 |
| chr2_+_79025696 | 0.03 |
ENST00000272324.10
|
REG3G
|
regenerating family member 3 gamma |
| chr15_+_22094522 | 0.03 |
ENST00000328795.5
|
OR4N4
|
olfactory receptor family 4 subfamily N member 4 |
| chr19_-_14835162 | 0.03 |
ENST00000322301.5
|
OR7A5
|
olfactory receptor family 7 subfamily A member 5 |
| chr8_-_37849838 | 0.03 |
ENST00000520601.5
ENST00000220659.11 ENST00000521170.5 |
BRF2
|
BRF2 RNA polymerase III transcription initiation factor subunit |
| chr1_+_35883189 | 0.03 |
ENST00000674304.1
ENST00000373204.6 ENST00000674426.1 |
AGO1
|
argonaute RISC component 1 |
| chr12_-_2876986 | 0.03 |
ENST00000342628.6
ENST00000361953.7 |
FOXM1
|
forkhead box M1 |
| chr6_-_33418046 | 0.03 |
ENST00000494751.5
ENST00000374496.3 |
CUTA
|
cutA divalent cation tolerance homolog |
| chr20_-_37095985 | 0.03 |
ENST00000344359.7
ENST00000373664.8 |
RBL1
|
RB transcriptional corepressor like 1 |
| chr2_-_36966503 | 0.03 |
ENST00000263918.9
|
STRN
|
striatin |
| chr18_-_27185284 | 0.03 |
ENST00000580774.2
ENST00000618847.5 |
CHST9
|
carbohydrate sulfotransferase 9 |
| chr6_-_75243770 | 0.02 |
ENST00000472311.6
ENST00000460985.1 ENST00000377978.3 ENST00000684430.1 ENST00000509698.6 |
COX7A2
|
cytochrome c oxidase subunit 7A2 |
| chr12_-_75209814 | 0.02 |
ENST00000549446.6
|
KCNC2
|
potassium voltage-gated channel subfamily C member 2 |
| chr9_-_134917872 | 0.02 |
ENST00000616356.4
ENST00000371806.4 |
FCN1
|
ficolin 1 |
| chr7_+_157336988 | 0.02 |
ENST00000262177.9
ENST00000417758.5 ENST00000443280.5 |
DNAJB6
|
DnaJ heat shock protein family (Hsp40) member B6 |
| chr7_-_64982021 | 0.02 |
ENST00000610793.1
ENST00000620222.4 |
ZNF117
|
zinc finger protein 117 |
| chr5_-_179623098 | 0.02 |
ENST00000630639.1
ENST00000329433.10 ENST00000510411.5 |
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 |
| chr6_-_31158073 | 0.02 |
ENST00000507751.5
ENST00000448162.6 ENST00000502557.5 ENST00000503420.5 ENST00000507892.1 ENST00000507226.1 ENST00000513222.1 ENST00000503934.5 ENST00000396263.6 ENST00000508683.5 ENST00000428174.1 ENST00000448141.6 ENST00000507829.5 ENST00000455279.6 ENST00000376266.9 |
CCHCR1
|
coiled-coil alpha-helical rod protein 1 |
| chr5_+_159263282 | 0.02 |
ENST00000296786.8
|
UBLCP1
|
ubiquitin like domain containing CTD phosphatase 1 |
| chr11_-_118176576 | 0.02 |
ENST00000278947.6
|
SCN2B
|
sodium voltage-gated channel beta subunit 2 |
| chrX_-_149505274 | 0.02 |
ENST00000428056.6
ENST00000340855.11 ENST00000370441.8 |
IDS
|
iduronate 2-sulfatase |
| chr5_-_56116946 | 0.02 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr1_-_12898270 | 0.02 |
ENST00000235347.4
|
PRAMEF10
|
PRAME family member 10 |
| chr2_+_44275457 | 0.02 |
ENST00000611973.4
ENST00000409387.5 |
SLC3A1
|
solute carrier family 3 member 1 |
| chr6_-_33418093 | 0.02 |
ENST00000488034.6
|
CUTA
|
cutA divalent cation tolerance homolog |
| chr5_-_147081462 | 0.02 |
ENST00000508267.5
ENST00000504198.5 |
PPP2R2B
|
protein phosphatase 2 regulatory subunit Bbeta |
| chr7_+_64903247 | 0.02 |
ENST00000476120.1
|
ZNF273
|
zinc finger protein 273 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX18
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.6 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 0.3 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.4 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.3 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.1 | GO:1903259 | exon-exon junction complex disassembly(GO:1903259) |
| 0.0 | 0.1 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.0 | 0.1 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.5 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 0.0 | 0.1 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.0 | 0.1 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.0 | 0.0 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 0.0 | 0.3 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.1 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.0 | 0.1 | GO:2001193 | gamma-delta T cell activation involved in immune response(GO:0002290) negative regulation of interferon-beta secretion(GO:0035548) regulation of gamma-delta T cell activation involved in immune response(GO:2001191) positive regulation of gamma-delta T cell activation involved in immune response(GO:2001193) |
| 0.0 | 0.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.1 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.1 | GO:0097460 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.0 | 0.1 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.0 | 0.2 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.4 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.1 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.1 | GO:1990822 | L-cystine transport(GO:0015811) basic amino acid transmembrane transport(GO:1990822) |
| 0.0 | 0.1 | GO:1901189 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.6 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 0.3 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.3 | GO:0004339 | glucan 1,4-alpha-glucosidase activity(GO:0004339) |
| 0.1 | 0.4 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.4 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.1 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.2 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.6 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.0 | GO:0051990 | (R)-2-hydroxyglutarate dehydrogenase activity(GO:0051990) |
| 0.0 | 0.1 | GO:0030107 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.0 | 0.1 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |