Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
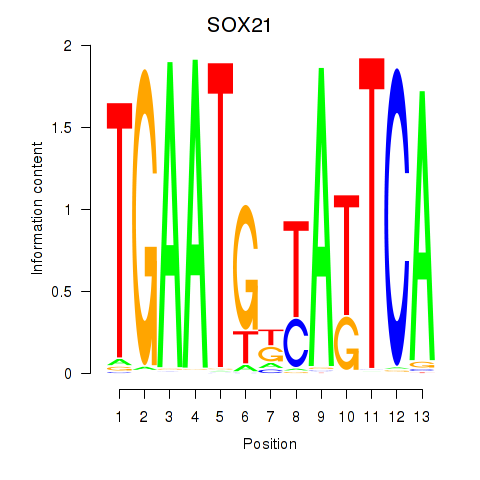
Results for SOX21
Z-value: 0.39
Transcription factors associated with SOX21
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX21
|
ENSG00000125285.6 | SRY-box transcription factor 21 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX21 | hg38_v1_chr13_-_94712505_94712553 | -0.03 | 9.4e-01 | Click! |
Activity profile of SOX21 motif
Sorted Z-values of SOX21 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_35180342 | 0.38 |
ENST00000639002.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr3_+_58008350 | 0.29 |
ENST00000490882.5
ENST00000358537.7 ENST00000429972.6 ENST00000682871.1 ENST00000295956.9 |
FLNB
|
filamin B |
| chr1_+_152908538 | 0.27 |
ENST00000368764.4
|
IVL
|
involucrin |
| chr8_+_100158576 | 0.27 |
ENST00000388798.7
|
SPAG1
|
sperm associated antigen 1 |
| chr11_+_118077009 | 0.24 |
ENST00000616579.4
ENST00000534111.5 |
TMPRSS4
|
transmembrane serine protease 4 |
| chr11_+_118077067 | 0.23 |
ENST00000522307.5
ENST00000523251.5 ENST00000437212.8 ENST00000522824.5 ENST00000522151.5 |
TMPRSS4
|
transmembrane serine protease 4 |
| chr8_-_86069662 | 0.23 |
ENST00000276616.3
|
PSKH2
|
protein serine kinase H2 |
| chr11_+_35180279 | 0.21 |
ENST00000531873.5
|
CD44
|
CD44 molecule (Indian blood group) |
| chr9_-_101487120 | 0.20 |
ENST00000374848.8
|
PGAP4
|
post-GPI attachment to proteins GalNAc transferase 4 |
| chr12_+_34022462 | 0.20 |
ENST00000538927.1
ENST00000266483.7 |
ALG10
|
ALG10 alpha-1,2-glucosyltransferase |
| chr4_+_68447453 | 0.18 |
ENST00000305363.9
|
TMPRSS11E
|
transmembrane serine protease 11E |
| chr6_+_116399395 | 0.18 |
ENST00000644499.1
|
ENSG00000285446.1
|
novel protein |
| chr6_+_31403553 | 0.17 |
ENST00000449934.7
ENST00000421350.1 |
MICA
|
MHC class I polypeptide-related sequence A |
| chrX_+_3066833 | 0.17 |
ENST00000359361.2
|
ARSF
|
arylsulfatase F |
| chr9_-_101487091 | 0.16 |
ENST00000374847.5
|
PGAP4
|
post-GPI attachment to proteins GalNAc transferase 4 |
| chrX_-_153944621 | 0.15 |
ENST00000393700.8
|
RENBP
|
renin binding protein |
| chr1_-_169485931 | 0.14 |
ENST00000367804.4
ENST00000646596.1 ENST00000236137.10 |
SLC19A2
|
solute carrier family 19 member 2 |
| chr11_+_89924064 | 0.13 |
ENST00000623787.3
|
TRIM49D2
|
tripartite motif containing 49D2 |
| chr12_+_122527229 | 0.13 |
ENST00000450485.6
ENST00000333479.12 |
KNTC1
|
kinetochore associated 1 |
| chr3_+_186570663 | 0.12 |
ENST00000265028.8
|
DNAJB11
|
DnaJ heat shock protein family (Hsp40) member B11 |
| chr4_+_74308463 | 0.12 |
ENST00000413830.6
|
EPGN
|
epithelial mitogen |
| chr14_+_21030201 | 0.11 |
ENST00000321760.11
ENST00000460647.6 ENST00000530140.6 ENST00000472458.5 |
TPPP2
|
tubulin polymerization promoting protein family member 2 |
| chrX_-_153944655 | 0.11 |
ENST00000369997.7
|
RENBP
|
renin binding protein |
| chr12_-_10998304 | 0.10 |
ENST00000538986.2
|
TAS2R20
|
taste 2 receptor member 20 |
| chr3_-_155854375 | 0.10 |
ENST00000643144.2
ENST00000359479.7 ENST00000646424.1 |
SLC33A1
|
solute carrier family 33 member 1 |
| chrX_+_66164210 | 0.10 |
ENST00000343002.7
ENST00000336279.9 |
HEPH
|
hephaestin |
| chr12_-_89352487 | 0.10 |
ENST00000548755.1
ENST00000279488.8 |
DUSP6
|
dual specificity phosphatase 6 |
| chr12_+_57591158 | 0.09 |
ENST00000422156.7
ENST00000354947.10 ENST00000540759.6 ENST00000551772.5 ENST00000550465.5 |
PIP4K2C
|
phosphatidylinositol-5-phosphate 4-kinase type 2 gamma |
| chr12_+_101280093 | 0.09 |
ENST00000261637.5
|
UTP20
|
UTP20 small subunit processome component |
| chr1_-_26067622 | 0.09 |
ENST00000374272.4
|
TRIM63
|
tripartite motif containing 63 |
| chr1_-_217076889 | 0.09 |
ENST00000493748.5
ENST00000463665.5 |
ESRRG
|
estrogen related receptor gamma |
| chr4_-_176269213 | 0.09 |
ENST00000296525.7
|
ASB5
|
ankyrin repeat and SOCS box containing 5 |
| chr14_+_22226711 | 0.09 |
ENST00000390463.3
|
TRAV36DV7
|
T cell receptor alpha variable 36/delta variable 7 |
| chr12_-_89352395 | 0.08 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6 |
| chr6_+_151494007 | 0.08 |
ENST00000239374.8
|
CCDC170
|
coiled-coil domain containing 170 |
| chr20_+_44715360 | 0.08 |
ENST00000190983.5
|
CCN5
|
cellular communication network factor 5 |
| chr8_-_141367276 | 0.08 |
ENST00000377741.4
|
GPR20
|
G protein-coupled receptor 20 |
| chr1_+_207325629 | 0.08 |
ENST00000618707.2
|
CD55
|
CD55 molecule (Cromer blood group) |
| chr11_+_5389377 | 0.08 |
ENST00000328611.5
|
OR51M1
|
olfactory receptor family 51 subfamily M member 1 |
| chr7_-_16421524 | 0.08 |
ENST00000407010.7
|
CRPPA
|
CDP-L-ribitol pyrophosphorylase A |
| chr7_+_2354810 | 0.07 |
ENST00000360876.9
ENST00000413917.5 ENST00000397011.2 |
EIF3B
|
eukaryotic translation initiation factor 3 subunit B |
| chr15_-_79971164 | 0.07 |
ENST00000335661.6
ENST00000267953.4 ENST00000677151.1 |
BCL2A1
|
BCL2 related protein A1 |
| chr10_-_12042771 | 0.07 |
ENST00000357604.10
|
UPF2
|
UPF2 regulator of nonsense mediated mRNA decay |
| chr5_+_176448363 | 0.07 |
ENST00000261942.7
|
FAF2
|
Fas associated factor family member 2 |
| chr16_-_18433412 | 0.07 |
ENST00000620440.4
|
NOMO2
|
NODAL modulator 2 |
| chr1_+_67166448 | 0.06 |
ENST00000347310.10
|
IL23R
|
interleukin 23 receptor |
| chr3_-_155806260 | 0.05 |
ENST00000534941.2
ENST00000340171.7 |
C3orf33
|
chromosome 3 open reading frame 33 |
| chr18_+_32190033 | 0.05 |
ENST00000269202.11
|
MEP1B
|
meprin A subunit beta |
| chr6_+_42563981 | 0.05 |
ENST00000372899.6
ENST00000372901.2 |
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chrX_-_101291325 | 0.05 |
ENST00000356784.2
|
TAF7L
|
TATA-box binding protein associated factor 7 like |
| chr18_+_32190015 | 0.05 |
ENST00000581447.1
|
MEP1B
|
meprin A subunit beta |
| chr1_+_200027605 | 0.05 |
ENST00000236914.7
|
NR5A2
|
nuclear receptor subfamily 5 group A member 2 |
| chr7_+_138076453 | 0.05 |
ENST00000242375.8
ENST00000411726.6 |
AKR1D1
|
aldo-keto reductase family 1 member D1 |
| chr14_+_103529163 | 0.04 |
ENST00000389749.9
|
TRMT61A
|
tRNA methyltransferase 61A |
| chr1_+_158931539 | 0.04 |
ENST00000368140.6
ENST00000368138.7 ENST00000392254.6 ENST00000392252.7 ENST00000368135.4 |
PYHIN1
|
pyrin and HIN domain family member 1 |
| chr14_-_106470788 | 0.03 |
ENST00000434710.1
|
IGHV3-43
|
immunoglobulin heavy variable 3-43 |
| chr8_+_127409026 | 0.03 |
ENST00000465342.4
|
POU5F1B
|
POU class 5 homeobox 1B |
| chr11_-_8263858 | 0.03 |
ENST00000534484.1
ENST00000335790.8 |
LMO1
|
LIM domain only 1 |
| chrX_+_131083706 | 0.03 |
ENST00000370921.1
|
ARHGAP36
|
Rho GTPase activating protein 36 |
| chr15_+_58410543 | 0.02 |
ENST00000356113.10
ENST00000414170.7 |
LIPC
|
lipase C, hepatic type |
| chr1_+_200027702 | 0.02 |
ENST00000367362.8
|
NR5A2
|
nuclear receptor subfamily 5 group A member 2 |
| chr10_-_13234329 | 0.02 |
ENST00000463405.2
|
UCMA
|
upper zone of growth plate and cartilage matrix associated |
| chr3_+_36993350 | 0.02 |
ENST00000536378.5
ENST00000673899.1 ENST00000231790.8 ENST00000673715.1 ENST00000456676.6 ENST00000673673.1 |
MLH1
|
mutL homolog 1 |
| chr3_-_36993103 | 0.02 |
ENST00000322716.8
|
EPM2AIP1
|
EPM2A interacting protein 1 |
| chr10_-_13234368 | 0.02 |
ENST00000378681.8
|
UCMA
|
upper zone of growth plate and cartilage matrix associated |
| chr6_+_131637296 | 0.01 |
ENST00000358229.6
ENST00000357639.8 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chrX_-_51682831 | 0.01 |
ENST00000634648.1
|
CENPVL2
|
centromere protein V like 2 |
| chr19_-_2783241 | 0.01 |
ENST00000676943.1
ENST00000589251.5 ENST00000221566.7 ENST00000676984.1 |
SGTA
|
small glutamine rich tetratricopeptide repeat containing alpha |
| chr6_+_29396555 | 0.01 |
ENST00000623183.1
|
OR12D2
|
olfactory receptor family 12 subfamily D member 2 |
| chr19_-_7040179 | 0.01 |
ENST00000381394.9
|
MBD3L4
|
methyl-CpG binding domain protein 3 like 4 |
| chr3_+_36993764 | 0.01 |
ENST00000674019.1
ENST00000458205.6 |
MLH1
|
mutL homolog 1 |
| chr14_+_92121953 | 0.00 |
ENST00000298875.9
ENST00000553427.5 |
CPSF2
|
cleavage and polyadenylation specific factor 2 |
| chr7_-_73719629 | 0.00 |
ENST00000395155.3
ENST00000395154.7 ENST00000395156.7 ENST00000222812.8 |
STX1A
|
syntaxin 1A |
| chr1_+_203795614 | 0.00 |
ENST00000367210.3
ENST00000432282.5 ENST00000453771.5 ENST00000367214.5 ENST00000639812.1 ENST00000367212.7 ENST00000332127.8 ENST00000550078.2 |
ZC3H11A
ZBED6
|
zinc finger CCCH-type containing 11A zinc finger BED-type containing 6 |
| chr4_-_103719980 | 0.00 |
ENST00000304883.3
|
TACR3
|
tachykinin receptor 3 |
| chrX_-_139642518 | 0.00 |
ENST00000370573.8
ENST00000338585.6 |
MCF2
|
MCF.2 cell line derived transforming sequence |
| chrX_+_11760035 | 0.00 |
ENST00000482871.6
ENST00000648013.1 |
MSL3
|
MSL complex subunit 3 |
| chr19_-_2783308 | 0.00 |
ENST00000677562.1
ENST00000677754.1 |
SGTA
|
small glutamine rich tetratricopeptide repeat containing alpha |
| chr22_+_22720615 | 0.00 |
ENST00000390309.2
|
IGLV3-19
|
immunoglobulin lambda variable 3-19 |
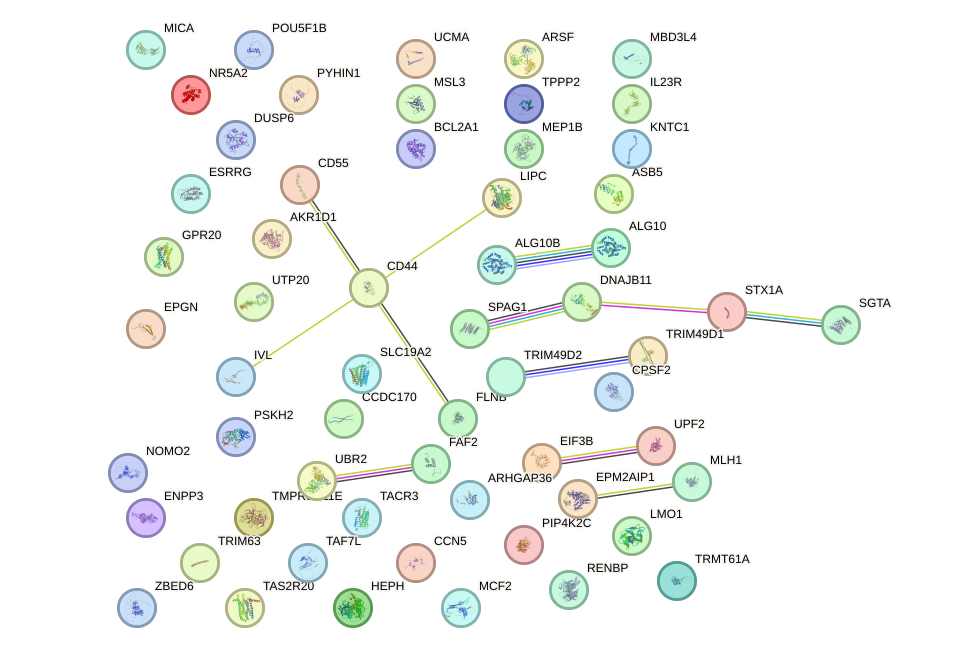
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX21
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.6 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.3 | GO:0006051 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.0 | 0.2 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.1 | GO:0000472 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.0 | 0.2 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.0 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.0 | 0.1 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.3 | GO:0007016 | keratinocyte development(GO:0003334) cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.0 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.0 | 0.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.2 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.3 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |