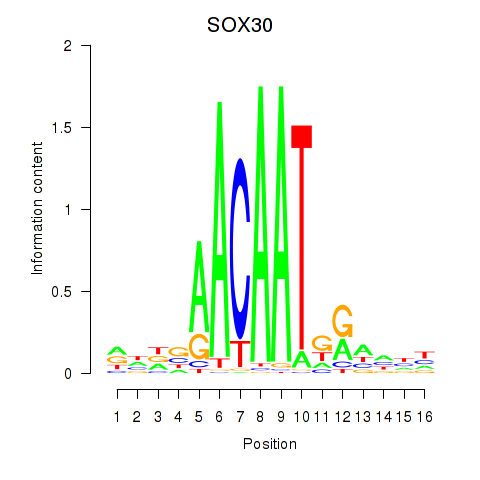
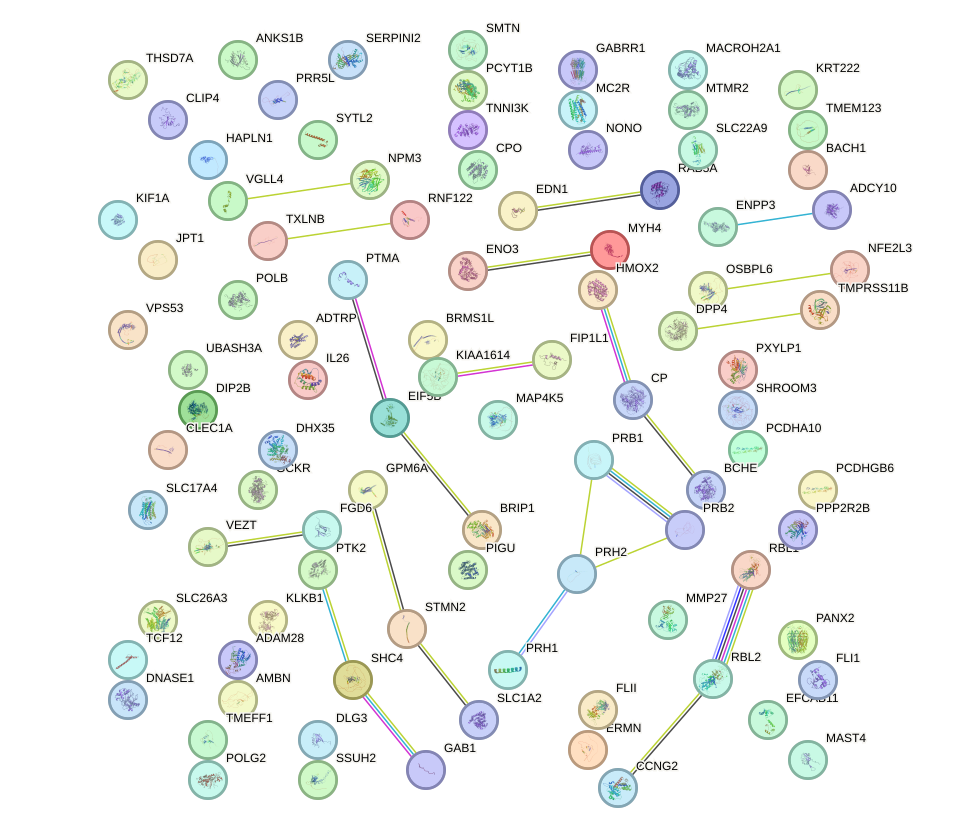
|
chr4_+_76435216
Show fit
|
0.22 |
ENST00000296043.7
|
SHROOM3
|
shroom family member 3
|
|
chr6_-_11778781
Show fit
|
0.20 |
ENST00000414691.8
ENST00000229583.9
|
ADTRP
|
androgen dependent TFPI regulating protein
|
|
chr5_-_83673544
Show fit
|
0.15 |
ENST00000503117.1
ENST00000510978.5
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1
|
|
chrX_-_24672654
Show fit
|
0.15 |
ENST00000379145.5
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr2_+_206939515
Show fit
|
0.15 |
ENST00000272852.4
|
CPO
|
carboxypeptidase O
|
|
chr11_+_128694052
Show fit
|
0.13 |
ENST00000527786.7
ENST00000534087.3
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor
|
|
chr13_-_85799400
Show fit
|
0.13 |
ENST00000647374.2
|
SLITRK6
|
SLIT and NTRK like family member 6
|
|
chr11_+_128693887
Show fit
|
0.13 |
ENST00000281428.12
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor
|
|
chr12_-_95217373
Show fit
|
0.13 |
ENST00000549499.1
ENST00000546711.5
ENST00000343958.9
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6
|
|
chrX_-_117973579
Show fit
|
0.11 |
ENST00000371878.5
|
KLHL13
|
kelch like family member 13
|
|
chrX_+_70444827
Show fit
|
0.11 |
ENST00000374360.8
|
DLG3
|
discs large MAGUK scaffold protein 3
|
|
chr4_+_70592295
Show fit
|
0.11 |
ENST00000449493.2
|
AMBN
|
ameloblastin
|
|
chr7_+_26152188
Show fit
|
0.10 |
ENST00000056233.4
|
NFE2L3
|
nuclear factor, erythroid 2 like 3
|
|
chr16_+_3654683
Show fit
|
0.10 |
ENST00000246949.10
|
DNASE1
|
deoxyribonuclease 1
|
|
chr11_+_63369779
Show fit
|
0.08 |
ENST00000279178.4
|
SLC22A9
|
solute carrier family 22 member 9
|
|
chr2_+_29113989
Show fit
|
0.08 |
ENST00000404424.5
|
CLIP4
|
CAP-Gly domain containing linker protein family member 4
|
|
chr5_+_140855882
Show fit
|
0.07 |
ENST00000562220.2
ENST00000307360.6
ENST00000506939.6
|
PCDHA10
|
protocadherin alpha 10
|
|
chr12_+_50504970
Show fit
|
0.07 |
ENST00000301180.10
|
DIP2B
|
disco interacting protein 2 homolog B
|
|
chr6_-_139291987
Show fit
|
0.06 |
ENST00000358430.8
|
TXLNB
|
taxilin beta
|
|
chr8_+_42338454
Show fit
|
0.06 |
ENST00000532157.5
ENST00000520008.5
|
POLB
|
DNA polymerase beta
|
|
chr11_-_85719160
Show fit
|
0.06 |
ENST00000389958.7
ENST00000527794.5
|
SYTL2
|
synaptotagmin like 2
|
|
chr16_+_4495852
Show fit
|
0.06 |
ENST00000575129.5
ENST00000398595.7
ENST00000414777.5
|
HMOX2
|
heme oxygenase 2
|
|
chr11_-_85719111
Show fit
|
0.05 |
ENST00000529581.5
ENST00000533577.1
|
SYTL2
|
synaptotagmin like 2
|
|
chrX_-_15854743
Show fit
|
0.05 |
ENST00000450644.2
|
AP1S2
|
adaptor related protein complex 1 subunit sigma 2
|
|
chr8_+_42338477
Show fit
|
0.05 |
ENST00000518925.5
ENST00000265421.9
|
POLB
|
DNA polymerase beta
|
|
chr8_+_79611036
Show fit
|
0.05 |
ENST00000220876.12
ENST00000518111.5
|
STMN2
|
stathmin 2
|
|
chr3_+_141262614
Show fit
|
0.05 |
ENST00000504264.5
|
PXYLP1
|
2-phosphoxylose phosphatase 1
|
|
chr6_+_12290353
Show fit
|
0.05 |
ENST00000379375.6
|
EDN1
|
endothelin 1
|
|
chr5_+_66958870
Show fit
|
0.04 |
ENST00000405643.5
ENST00000407621.1
ENST00000432426.5
|
MAST4
|
microtubule associated serine/threonine kinase family member 4
|
|
chr9_+_100473140
Show fit
|
0.04 |
ENST00000374879.5
|
TMEFF1
|
transmembrane protein with EGF like and two follistatin like domains 1
|
|
chr2_+_27496830
Show fit
|
0.04 |
ENST00000264717.7
|
GCKR
|
glucokinase regulator
|
|
chr4_+_143381939
Show fit
|
0.04 |
ENST00000505913.5
|
GAB1
|
GRB2 associated binding protein 1
|
|
chr6_+_131637296
Show fit
|
0.04 |
ENST00000358229.6
ENST00000357639.8
|
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3
|
|
chr22_+_31092447
Show fit
|
0.04 |
ENST00000455608.5
|
SMTN
|
smoothelin
|
|
chr7_+_142670734
Show fit
|
0.04 |
ENST00000390398.3
|
TRBV25-1
|
T cell receptor beta variable 25-1
|
|
chr4_+_186227501
Show fit
|
0.04 |
ENST00000446598.6
ENST00000264690.11
ENST00000513864.2
|
KLKB1
|
kallikrein B1
|
|
chr11_-_35419899
Show fit
|
0.04 |
ENST00000646847.1
ENST00000449068.2
ENST00000643401.1
ENST00000645966.1
ENST00000647104.1
|
SLC1A2
|
solute carrier family 1 member 2
|
|
chr8_-_140800535
Show fit
|
0.04 |
ENST00000521986.5
ENST00000523539.5
|
PTK2
|
protein tyrosine kinase 2
|
|
chr17_-_40665121
Show fit
|
0.04 |
ENST00000394052.5
|
KRT222
|
keratin 222
|
|
chr8_-_33567118
Show fit
|
0.03 |
ENST00000256257.2
|
RNF122
|
ring finger protein 122
|
|
chr11_-_85719045
Show fit
|
0.03 |
ENST00000533057.6
ENST00000533892.5
|
SYTL2
|
synaptotagmin like 2
|
|
chr4_+_53377630
Show fit
|
0.03 |
ENST00000337488.11
ENST00000358575.9
ENST00000507922.5
ENST00000306932.10
|
FIP1L1
|
factor interacting with PAPOLA and CPSF1
|
|
chr1_+_180928133
Show fit
|
0.03 |
ENST00000367587.1
|
KIAA1614
|
KIAA1614
|
|
chr4_+_77157189
Show fit
|
0.03 |
ENST00000316355.10
ENST00000502280.5
|
CCNG2
|
cyclin G2
|
|
chr2_+_231708511
Show fit
|
0.03 |
ENST00000341369.11
ENST00000409115.8
ENST00000409683.5
|
PTMA
|
prothymosin alpha
|
|
chr12_-_10098977
Show fit
|
0.03 |
ENST00000315330.8
ENST00000457018.6
|
CLEC1A
|
C-type lectin domain family 1 member A
|
|
chrX_+_41333342
Show fit
|
0.03 |
ENST00000629496.3
ENST00000625837.2
ENST00000626301.2
|
DDX3X
|
DEAD-box helicase 3 X-linked
|
|
chr4_-_68245683
Show fit
|
0.03 |
ENST00000332644.6
|
TMPRSS11B
|
transmembrane serine protease 11B
|
|
chr3_-_8644782
Show fit
|
0.03 |
ENST00000544814.6
|
SSUH2
|
ssu-2 homolog
|
|
chr7_-_149028452
Show fit
|
0.03 |
ENST00000413966.1
ENST00000652332.1
|
PDIA4
|
protein disulfide isomerase family A member 4
|
|
chr7_-_11832190
Show fit
|
0.03 |
ENST00000423059.9
ENST00000617773.1
|
THSD7A
|
thrombospondin type 1 domain containing 7A
|
|
chr11_-_35419213
Show fit
|
0.03 |
ENST00000642171.1
ENST00000644050.1
ENST00000643134.1
|
SLC1A2
|
solute carrier family 1 member 2
|
|
chr11_-_35420017
Show fit
|
0.02 |
ENST00000643000.1
ENST00000646099.1
ENST00000647372.1
ENST00000642578.1
|
SLC1A2
|
solute carrier family 1 member 2
|
|
chr4_+_53377749
Show fit
|
0.02 |
ENST00000507166.5
|
ENSG00000282278.1
|
novel FIP1L1-PDGFRA fusion protein
|
|
chr6_-_127900958
Show fit
|
0.02 |
ENST00000434358.3
ENST00000630369.2
ENST00000368248.4
|
THEMIS
|
thymocyte selection associated
|
|
chr18_-_13915531
Show fit
|
0.02 |
ENST00000327606.4
|
MC2R
|
melanocortin 2 receptor
|
|
chr12_+_95217792
Show fit
|
0.02 |
ENST00000436874.6
ENST00000551472.5
ENST00000552821.5
|
VEZT
|
vezatin, adherens junctions transmembrane protein
|
|
chr11_-_35420050
Show fit
|
0.02 |
ENST00000395753.6
ENST00000395750.6
ENST00000645634.1
|
SLC1A2
|
solute carrier family 1 member 2
|
|
chr3_-_167474026
Show fit
|
0.02 |
ENST00000466903.1
ENST00000264677.8
|
SERPINI2
|
serpin family I member 2
|
|
chr17_-_10469558
Show fit
|
0.02 |
ENST00000255381.2
|
MYH4
|
myosin heavy chain 4
|
|
chr2_+_99337364
Show fit
|
0.02 |
ENST00000617677.1
ENST00000289371.11
|
EIF5B
|
eukaryotic translation initiation factor 5B
|
|
chr11_-_102452758
Show fit
|
0.02 |
ENST00000398136.7
ENST00000361236.7
|
TMEM123
|
transmembrane protein 123
|
|
chr11_-_35419462
Show fit
|
0.02 |
ENST00000643522.1
|
SLC1A2
|
solute carrier family 1 member 2
|
|
chr1_-_167914089
Show fit
|
0.02 |
ENST00000476818.2
ENST00000367848.1
ENST00000367851.9
ENST00000545172.5
|
ADCY10
|
adenylate cyclase 10
|
|
chr3_-_149221811
Show fit
|
0.02 |
ENST00000455472.3
ENST00000264613.11
|
CP
|
ceruloplasmin
|
|
chr17_-_676348
Show fit
|
0.02 |
ENST00000681510.1
ENST00000679680.1
|
VPS53
|
VPS53 subunit of GARP complex
|
|
chr5_-_135399863
Show fit
|
0.01 |
ENST00000510038.1
ENST00000304332.8
|
MACROH2A1
|
macroH2A.1 histone
|
|
chr8_+_24294107
Show fit
|
0.01 |
ENST00000437154.6
|
ADAM28
|
ADAM metallopeptidase domain 28
|
|
chrX_+_71283186
Show fit
|
0.01 |
ENST00000535149.5
|
NONO
|
non-POU domain containing octamer binding
|
|
chr15_-_48963912
Show fit
|
0.01 |
ENST00000332408.9
|
SHC4
|
SHC adaptor protein 4
|
|
chr11_-_95910748
Show fit
|
0.01 |
ENST00000675933.1
|
MTMR2
|
myotubularin related protein 2
|
|
chr5_+_141408000
Show fit
|
0.01 |
ENST00000616430.1
|
PCDHGB6
|
protocadherin gamma subfamily B, 6
|
|
chr2_-_240820945
Show fit
|
0.01 |
ENST00000428768.2
ENST00000650053.1
ENST00000650130.1
|
KIF1A
|
kinesin family member 1A
|
|
chr5_-_147081428
Show fit
|
0.01 |
ENST00000394413.7
|
PPP2R2B
|
protein phosphatase 2 regulatory subunit Bbeta
|
|
chr14_+_35826298
Show fit
|
0.01 |
ENST00000216807.12
|
BRMS1L
|
BRMS1 like transcriptional repressor
|
|
chr2_-_157325808
Show fit
|
0.01 |
ENST00000410096.6
ENST00000420719.6
ENST00000409216.5
ENST00000419116.2
|
ERMN
|
ermin
|
|
chr17_+_4948252
Show fit
|
0.01 |
ENST00000520221.5
|
ENO3
|
enolase 3
|
|
chr2_+_178320228
Show fit
|
0.01 |
ENST00000315022.2
|
OSBPL6
|
oxysterol binding protein like 6
|
|
chr1_+_74235377
Show fit
|
0.01 |
ENST00000326637.8
|
TNNI3K
|
TNNI3 interacting kinase
|
|
chr12_-_98894830
Show fit
|
0.01 |
ENST00000549797.5
ENST00000333732.11
ENST00000341752.11
|
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr15_+_56918612
Show fit
|
0.01 |
ENST00000438423.6
ENST00000267811.9
ENST00000333725.10
ENST00000559609.5
|
TCF12
|
transcription factor 12
|
|
chr22_+_50170720
Show fit
|
0.01 |
ENST00000159647.9
ENST00000395842.3
|
PANX2
|
pannexin 2
|
|
chr7_-_107803215
Show fit
|
0.01 |
ENST00000340010.10
ENST00000453332.1
|
SLC26A3
|
solute carrier family 26 member 3
|
|
chr11_-_95910665
Show fit
|
0.01 |
ENST00000674610.1
ENST00000675660.1
|
MTMR2
|
myotubularin related protein 2
|
|
chr6_+_25754699
Show fit
|
0.01 |
ENST00000439485.6
ENST00000377905.9
|
SLC17A4
|
solute carrier family 17 member 4
|
|
chr3_-_11643871
Show fit
|
0.01 |
ENST00000430365.7
|
VGLL4
|
vestigial like family member 4
|
|
chr4_-_175812746
Show fit
|
0.00 |
ENST00000393658.6
|
GPM6A
|
glycoprotein M6A
|
|
chr20_+_38962299
Show fit
|
0.00 |
ENST00000373325.6
ENST00000373323.8
ENST00000252011.8
ENST00000615559.1
|
DHX35
ENSG00000277581.1
|
DEAH-box helicase 35
novel transcript, sense intronic to DHX35
|
|
chr11_+_36296281
Show fit
|
0.00 |
ENST00000530639.6
|
PRR5L
|
proline rich 5 like
|
|
chr8_+_24294044
Show fit
|
0.00 |
ENST00000265769.9
|
ADAM28
|
ADAM metallopeptidase domain 28
|
|
chrX_+_71283577
Show fit
|
0.00 |
ENST00000420903.6
ENST00000373856.8
ENST00000678437.1
ENST00000678830.1
ENST00000677879.1
ENST00000373841.5
ENST00000276079.13
ENST00000676797.1
ENST00000678660.1
ENST00000678231.1
ENST00000677612.1
ENST00000413858.5
ENST00000450092.6
|
NONO
|
non-POU domain containing octamer binding
|
|
chr3_-_165837412
Show fit
|
0.00 |
ENST00000479451.5
ENST00000488954.1
ENST00000264381.8
|
BCHE
|
butyrylcholinesterase
|
|
chr17_-_75153826
Show fit
|
0.00 |
ENST00000481647.5
ENST00000470924.5
|
JPT1
|
Jupiter microtubule associated homolog 1
|
|
chr12_-_68225806
Show fit
|
0.00 |
ENST00000229134.5
|
IL26
|
interleukin 26
|
|
chr3_+_19947074
Show fit
|
0.00 |
ENST00000273047.9
|
RAB5A
|
RAB5A, member RAS oncogene family
|
|
chr2_-_162074394
Show fit
|
0.00 |
ENST00000676810.1
|
DPP4
|
dipeptidyl peptidase 4
|
|
chr11_-_102705737
Show fit
|
0.00 |
ENST00000260229.5
|
MMP27
|
matrix metallopeptidase 27
|
|
chr14_-_89954518
Show fit
|
0.00 |
ENST00000556005.1
ENST00000555872.5
|
EFCAB11
|
EF-hand calcium binding domain 11
|
|
chr12_+_10929229
Show fit
|
0.00 |
ENST00000381847.7
ENST00000396400.4
|
PRH2
|
proline rich protein HaeIII subfamily 2
|
|
chr6_-_89217339
Show fit
|
0.00 |
ENST00000454853.7
|
GABRR1
|
gamma-aminobutyric acid type A receptor subunit rho1
|
|
chr1_-_118989504
Show fit
|
0.00 |
ENST00000207157.7
|
TBX15
|
T-box transcription factor 15
|
|
chr21_+_29299368
Show fit
|
0.00 |
ENST00000399921.5
|
BACH1
|
BTB domain and CNC homolog 1
|
|
chr12_-_11395556
Show fit
|
0.00 |
ENST00000565533.1
ENST00000389362.6
ENST00000546254.3
|
PRB2
PRB1
|
proline rich protein BstNI subfamily 2
proline rich protein BstNI subfamily 1
|
|
chr1_-_23369813
Show fit
|
0.00 |
ENST00000314011.9
|
ZNF436
|
zinc finger protein 436
|