Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
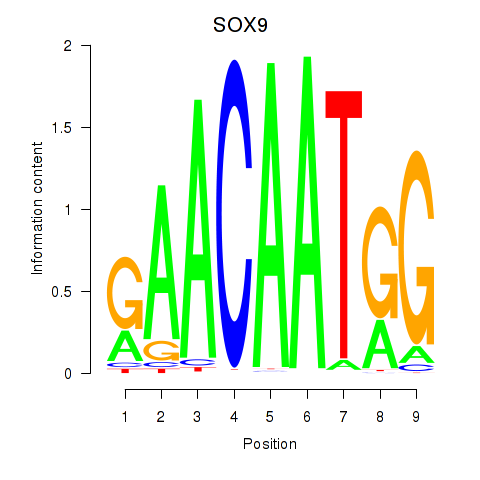
Results for SOX9
Z-value: 0.13
Transcription factors associated with SOX9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX9
|
ENSG00000125398.8 | SRY-box transcription factor 9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX9 | hg38_v1_chr17_+_72121012_72121033 | -0.16 | 7.0e-01 | Click! |
Activity profile of SOX9 motif
Sorted Z-values of SOX9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_121873089 | 0.33 |
ENST00000651065.1
|
PTPRZ1
|
protein tyrosine phosphatase receptor type Z1 |
| chr7_+_121873152 | 0.30 |
ENST00000650826.1
ENST00000650728.1 ENST00000393386.7 ENST00000651390.1 ENST00000651842.1 ENST00000650681.1 |
PTPRZ1
|
protein tyrosine phosphatase receptor type Z1 |
| chr9_+_2622053 | 0.26 |
ENST00000681306.1
ENST00000681618.1 |
VLDLR
|
very low density lipoprotein receptor |
| chr10_-_104085847 | 0.24 |
ENST00000648076.2
|
COL17A1
|
collagen type XVII alpha 1 chain |
| chrX_-_117973579 | 0.23 |
ENST00000371878.5
|
KLHL13
|
kelch like family member 13 |
| chr12_-_52493250 | 0.22 |
ENST00000330722.7
|
KRT6A
|
keratin 6A |
| chr16_+_788614 | 0.20 |
ENST00000262315.14
ENST00000455171.6 ENST00000317063.10 |
CHTF18
|
chromosome transmission fidelity factor 18 |
| chr8_+_94641074 | 0.19 |
ENST00000423620.6
|
ESRP1
|
epithelial splicing regulatory protein 1 |
| chr12_-_24902243 | 0.19 |
ENST00000538118.5
|
BCAT1
|
branched chain amino acid transaminase 1 |
| chr4_+_112860981 | 0.18 |
ENST00000671704.1
|
ANK2
|
ankyrin 2 |
| chr4_+_112860912 | 0.18 |
ENST00000671951.1
|
ANK2
|
ankyrin 2 |
| chr4_+_112861053 | 0.18 |
ENST00000672221.1
|
ANK2
|
ankyrin 2 |
| chr6_-_106975452 | 0.17 |
ENST00000619869.1
ENST00000619133.4 |
CD24
|
CD24 molecule |
| chr3_-_8644782 | 0.17 |
ENST00000544814.6
|
SSUH2
|
ssu-2 homolog |
| chr12_-_8662619 | 0.17 |
ENST00000544889.1
ENST00000543369.5 |
MFAP5
|
microfibril associated protein 5 |
| chr20_+_20368096 | 0.16 |
ENST00000310227.3
|
INSM1
|
INSM transcriptional repressor 1 |
| chr12_-_8662703 | 0.16 |
ENST00000535336.5
|
MFAP5
|
microfibril associated protein 5 |
| chr6_-_106975309 | 0.16 |
ENST00000615659.1
|
CD24
|
CD24 molecule |
| chr6_-_106974721 | 0.15 |
ENST00000606017.2
ENST00000620405.1 |
CD24
|
CD24 molecule |
| chr3_+_189789643 | 0.15 |
ENST00000354600.10
|
TP63
|
tumor protein p63 |
| chr12_+_77830886 | 0.14 |
ENST00000397909.7
ENST00000549464.5 |
NAV3
|
neuron navigator 3 |
| chr9_+_78297143 | 0.14 |
ENST00000347159.6
|
PSAT1
|
phosphoserine aminotransferase 1 |
| chr9_-_6645712 | 0.13 |
ENST00000321612.8
|
GLDC
|
glycine decarboxylase |
| chr12_-_95217373 | 0.13 |
ENST00000549499.1
ENST00000546711.5 ENST00000343958.9 |
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr5_+_140834230 | 0.13 |
ENST00000356878.5
ENST00000525929.2 |
PCDHA7
|
protocadherin alpha 7 |
| chr9_+_78297117 | 0.13 |
ENST00000376588.4
|
PSAT1
|
phosphoserine aminotransferase 1 |
| chr14_-_70809494 | 0.13 |
ENST00000381250.8
ENST00000554752.7 ENST00000555993.6 |
MAP3K9
|
mitogen-activated protein kinase kinase kinase 9 |
| chr8_-_80080816 | 0.13 |
ENST00000520527.5
ENST00000517427.5 ENST00000379097.7 ENST00000448733.3 |
TPD52
|
tumor protein D52 |
| chr22_+_21632751 | 0.12 |
ENST00000292779.4
|
CCDC116
|
coiled-coil domain containing 116 |
| chr5_-_16936231 | 0.12 |
ENST00000507288.1
ENST00000274203.13 ENST00000513610.6 |
MYO10
|
myosin X |
| chr9_+_27109393 | 0.12 |
ENST00000406359.8
|
TEK
|
TEK receptor tyrosine kinase |
| chr8_+_103819244 | 0.12 |
ENST00000262231.14
ENST00000507740.5 ENST00000408894.6 |
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr18_+_22169580 | 0.12 |
ENST00000269216.10
|
GATA6
|
GATA binding protein 6 |
| chr6_+_33620329 | 0.11 |
ENST00000374316.9
|
ITPR3
|
inositol 1,4,5-trisphosphate receptor type 3 |
| chr12_-_91058016 | 0.11 |
ENST00000266719.4
|
KERA
|
keratocan |
| chr15_+_40844018 | 0.11 |
ENST00000344051.8
ENST00000562057.6 |
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr8_+_101492423 | 0.10 |
ENST00000521085.1
ENST00000646743.1 |
GRHL2
|
grainyhead like transcription factor 2 |
| chr10_-_102114935 | 0.10 |
ENST00000361198.9
|
LDB1
|
LIM domain binding 1 |
| chr19_+_1249870 | 0.10 |
ENST00000591446.6
|
MIDN
|
midnolin |
| chr15_-_34367045 | 0.10 |
ENST00000617710.4
|
LPCAT4
|
lysophosphatidylcholine acyltransferase 4 |
| chr9_-_122227525 | 0.10 |
ENST00000373755.6
ENST00000373754.6 |
LHX6
|
LIM homeobox 6 |
| chr15_-_34367159 | 0.10 |
ENST00000314891.11
|
LPCAT4
|
lysophosphatidylcholine acyltransferase 4 |
| chr17_-_39607876 | 0.10 |
ENST00000302584.5
|
NEUROD2
|
neuronal differentiation 2 |
| chr3_+_189789734 | 0.09 |
ENST00000437221.5
ENST00000392463.6 ENST00000392461.7 ENST00000449992.5 ENST00000456148.1 |
TP63
|
tumor protein p63 |
| chr1_+_156149657 | 0.09 |
ENST00000414683.5
|
SEMA4A
|
semaphorin 4A |
| chr3_-_65597886 | 0.09 |
ENST00000460329.6
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr1_+_61952036 | 0.09 |
ENST00000646453.1
ENST00000635137.1 |
PATJ
|
PATJ crumbs cell polarity complex component |
| chr2_-_61538180 | 0.09 |
ENST00000677150.1
ENST00000678182.1 ENST00000677928.1 ENST00000406957.5 |
XPO1
|
exportin 1 |
| chr3_-_149333407 | 0.09 |
ENST00000470080.5
|
TM4SF18
|
transmembrane 4 L six family member 18 |
| chr11_-_79441016 | 0.08 |
ENST00000278550.12
|
TENM4
|
teneurin transmembrane protein 4 |
| chr8_-_109680812 | 0.08 |
ENST00000528716.5
ENST00000527600.5 ENST00000531230.5 ENST00000532189.5 ENST00000534184.5 ENST00000408889.7 ENST00000533171.5 |
SYBU
|
syntabulin |
| chr4_+_20251896 | 0.08 |
ENST00000504154.6
|
SLIT2
|
slit guidance ligand 2 |
| chr15_+_85380625 | 0.08 |
ENST00000560302.5
|
AKAP13
|
A-kinase anchoring protein 13 |
| chr22_-_46537593 | 0.08 |
ENST00000262738.9
ENST00000674500.2 |
CELSR1
|
cadherin EGF LAG seven-pass G-type receptor 1 |
| chr10_+_89701580 | 0.08 |
ENST00000371728.8
ENST00000260753.8 |
KIF20B
|
kinesin family member 20B |
| chr3_-_123620496 | 0.08 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr2_-_61538313 | 0.08 |
ENST00000677803.1
ENST00000677239.1 ENST00000401558.7 |
XPO1
|
exportin 1 |
| chr1_+_160400543 | 0.08 |
ENST00000368061.3
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr2_-_61538290 | 0.08 |
ENST00000678790.1
|
XPO1
|
exportin 1 |
| chr19_+_44891206 | 0.08 |
ENST00000405636.6
ENST00000252487.9 ENST00000592434.5 ENST00000589649.1 ENST00000426677.7 |
TOMM40
|
translocase of outer mitochondrial membrane 40 |
| chr3_-_123620571 | 0.08 |
ENST00000583087.5
|
MYLK
|
myosin light chain kinase |
| chr8_-_42501224 | 0.08 |
ENST00000520262.6
ENST00000517366.1 |
SLC20A2
|
solute carrier family 20 member 2 |
| chr12_-_85836372 | 0.08 |
ENST00000361228.5
|
RASSF9
|
Ras association domain family member 9 |
| chr12_-_10098940 | 0.08 |
ENST00000420265.2
|
CLEC1A
|
C-type lectin domain family 1 member A |
| chr1_+_26177482 | 0.08 |
ENST00000361530.11
ENST00000374253.9 |
CNKSR1
|
connector enhancer of kinase suppressor of Ras 1 |
| chr17_+_17042433 | 0.08 |
ENST00000651222.2
|
MPRIP
|
myosin phosphatase Rho interacting protein |
| chr4_+_53377749 | 0.08 |
ENST00000507166.5
|
ENSG00000282278.1
|
novel FIP1L1-PDGFRA fusion protein |
| chr19_+_10654327 | 0.08 |
ENST00000407004.7
ENST00000589998.5 ENST00000589600.5 |
ILF3
|
interleukin enhancer binding factor 3 |
| chr11_-_46846233 | 0.07 |
ENST00000529230.6
ENST00000312055.9 |
CKAP5
|
cytoskeleton associated protein 5 |
| chr7_-_11832190 | 0.07 |
ENST00000423059.9
ENST00000617773.1 |
THSD7A
|
thrombospondin type 1 domain containing 7A |
| chr2_-_61538516 | 0.07 |
ENST00000676771.1
ENST00000677814.1 ENST00000443240.5 ENST00000677556.1 ENST00000676553.1 |
XPO1
|
exportin 1 |
| chr7_+_77538059 | 0.07 |
ENST00000435495.6
|
PTPN12
|
protein tyrosine phosphatase non-receptor type 12 |
| chrX_+_68829009 | 0.07 |
ENST00000204961.5
|
EFNB1
|
ephrin B1 |
| chr6_-_85644043 | 0.07 |
ENST00000678930.1
ENST00000678355.1 |
SYNCRIP
|
synaptotagmin binding cytoplasmic RNA interacting protein |
| chr7_+_26152188 | 0.07 |
ENST00000056233.4
|
NFE2L3
|
nuclear factor, erythroid 2 like 3 |
| chr2_-_53970985 | 0.07 |
ENST00000404125.6
|
PSME4
|
proteasome activator subunit 4 |
| chrX_-_24672654 | 0.07 |
ENST00000379145.5
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr1_+_156150008 | 0.07 |
ENST00000355014.6
|
SEMA4A
|
semaphorin 4A |
| chr3_+_14402576 | 0.07 |
ENST00000613060.4
|
SLC6A6
|
solute carrier family 6 member 6 |
| chr11_+_73950985 | 0.07 |
ENST00000339764.6
|
DNAJB13
|
DnaJ heat shock protein family (Hsp40) member B13 |
| chr3_+_39467672 | 0.07 |
ENST00000436143.6
ENST00000441980.6 ENST00000682069.1 ENST00000311042.10 |
MOBP
|
myelin associated oligodendrocyte basic protein |
| chr12_-_10098977 | 0.07 |
ENST00000315330.8
ENST00000457018.6 |
CLEC1A
|
C-type lectin domain family 1 member A |
| chr1_+_147902789 | 0.07 |
ENST00000369235.2
|
GJA8
|
gap junction protein alpha 8 |
| chr1_-_243255170 | 0.07 |
ENST00000366542.6
|
CEP170
|
centrosomal protein 170 |
| chr19_+_10654261 | 0.07 |
ENST00000449870.5
|
ILF3
|
interleukin enhancer binding factor 3 |
| chr7_-_22193728 | 0.07 |
ENST00000620335.4
|
RAPGEF5
|
Rap guanine nucleotide exchange factor 5 |
| chr12_-_122500520 | 0.07 |
ENST00000540586.1
ENST00000543897.5 |
ZCCHC8
|
zinc finger CCHC-type containing 8 |
| chr5_+_138352674 | 0.07 |
ENST00000314358.10
|
KDM3B
|
lysine demethylase 3B |
| chr1_-_243255320 | 0.07 |
ENST00000366544.5
ENST00000366543.5 |
CEP170
|
centrosomal protein 170 |
| chr11_+_1390273 | 0.07 |
ENST00000526678.5
|
BRSK2
|
BR serine/threonine kinase 2 |
| chr7_+_107470050 | 0.07 |
ENST00000304402.6
|
GPR22
|
G protein-coupled receptor 22 |
| chr18_+_58863580 | 0.06 |
ENST00000586085.5
ENST00000589288.5 |
ZNF532
|
zinc finger protein 532 |
| chr19_+_55376818 | 0.06 |
ENST00000291934.4
|
TMEM190
|
transmembrane protein 190 |
| chr7_-_75738930 | 0.06 |
ENST00000336926.11
ENST00000434438.6 |
HIP1
|
huntingtin interacting protein 1 |
| chr2_+_54456311 | 0.06 |
ENST00000615901.4
ENST00000356805.9 |
SPTBN1
|
spectrin beta, non-erythrocytic 1 |
| chr20_-_51768327 | 0.06 |
ENST00000311637.9
ENST00000338821.6 |
ATP9A
|
ATPase phospholipid transporting 9A (putative) |
| chr6_+_32164586 | 0.06 |
ENST00000333845.11
ENST00000395512.5 ENST00000432129.1 |
EGFL8
|
EGF like domain multiple 8 |
| chr12_-_122500924 | 0.06 |
ENST00000633063.3
|
ZCCHC8
|
zinc finger CCHC-type containing 8 |
| chr7_-_22193824 | 0.06 |
ENST00000401957.6
|
RAPGEF5
|
Rap guanine nucleotide exchange factor 5 |
| chr9_+_22446808 | 0.06 |
ENST00000325870.3
|
DMRTA1
|
DMRT like family A1 |
| chr7_+_116672357 | 0.06 |
ENST00000456159.1
|
MET
|
MET proto-oncogene, receptor tyrosine kinase |
| chr8_+_24294107 | 0.06 |
ENST00000437154.6
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr2_-_157325808 | 0.06 |
ENST00000410096.6
ENST00000420719.6 ENST00000409216.5 ENST00000419116.2 |
ERMN
|
ermin |
| chr12_-_8662881 | 0.06 |
ENST00000433590.6
|
MFAP5
|
microfibril associated protein 5 |
| chr21_-_17819386 | 0.06 |
ENST00000400559.7
ENST00000400558.7 |
C21orf91
|
chromosome 21 open reading frame 91 |
| chr1_+_180928133 | 0.06 |
ENST00000367587.1
|
KIAA1614
|
KIAA1614 |
| chr3_+_39467598 | 0.06 |
ENST00000428261.5
ENST00000420739.5 ENST00000415443.5 ENST00000447324.5 ENST00000383754.7 |
MOBP
|
myelin associated oligodendrocyte basic protein |
| chr1_+_61952283 | 0.06 |
ENST00000307297.8
|
PATJ
|
PATJ crumbs cell polarity complex component |
| chr12_-_122500947 | 0.06 |
ENST00000672018.1
|
ZCCHC8
|
zinc finger CCHC-type containing 8 |
| chr16_+_3654683 | 0.06 |
ENST00000246949.10
|
DNASE1
|
deoxyribonuclease 1 |
| chr1_-_225427897 | 0.06 |
ENST00000421383.1
ENST00000272163.9 |
LBR
|
lamin B receptor |
| chr17_+_59155726 | 0.06 |
ENST00000578777.5
ENST00000577457.1 ENST00000582995.5 ENST00000262293.9 ENST00000614081.1 |
PRR11
|
proline rich 11 |
| chr15_+_40594241 | 0.06 |
ENST00000532056.5
ENST00000527044.5 ENST00000399668.7 |
KNL1
|
kinetochore scaffold 1 |
| chr4_+_53377630 | 0.06 |
ENST00000337488.11
ENST00000358575.9 ENST00000507922.5 ENST00000306932.10 |
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr16_+_30923565 | 0.06 |
ENST00000338343.10
|
FBXL19
|
F-box and leucine rich repeat protein 19 |
| chr15_+_40594001 | 0.06 |
ENST00000346991.9
ENST00000528975.5 |
KNL1
|
kinetochore scaffold 1 |
| chr3_+_111998915 | 0.06 |
ENST00000478951.6
|
TAGLN3
|
transgelin 3 |
| chr6_-_85643832 | 0.06 |
ENST00000677771.1
ENST00000676688.1 |
SYNCRIP
|
synaptotagmin binding cytoplasmic RNA interacting protein |
| chr1_-_16212598 | 0.06 |
ENST00000270747.8
|
ARHGEF19
|
Rho guanine nucleotide exchange factor 19 |
| chr3_+_14402592 | 0.06 |
ENST00000622186.5
ENST00000621751.4 |
SLC6A6
|
solute carrier family 6 member 6 |
| chr8_+_97644164 | 0.06 |
ENST00000336273.8
|
MTDH
|
metadherin |
| chr7_-_93226449 | 0.06 |
ENST00000394468.7
ENST00000453812.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr9_+_121699328 | 0.06 |
ENST00000373782.7
|
DAB2IP
|
DAB2 interacting protein |
| chr2_-_200963633 | 0.06 |
ENST00000234296.7
|
ORC2
|
origin recognition complex subunit 2 |
| chr11_-_116837586 | 0.06 |
ENST00000375320.5
ENST00000359492.6 ENST00000375329.6 ENST00000375323.5 ENST00000236850.5 |
APOA1
|
apolipoprotein A1 |
| chr16_+_89923333 | 0.06 |
ENST00000315491.12
ENST00000555576.5 ENST00000554336.5 ENST00000553967.1 |
TUBB3
|
tubulin beta 3 class III |
| chr1_-_182672232 | 0.05 |
ENST00000508450.5
|
RGS8
|
regulator of G protein signaling 8 |
| chr3_+_111999189 | 0.05 |
ENST00000455401.6
|
TAGLN3
|
transgelin 3 |
| chrX_-_130903187 | 0.05 |
ENST00000432489.5
ENST00000394363.6 ENST00000338144.8 |
ENOX2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr14_+_74763308 | 0.05 |
ENST00000325680.12
ENST00000552421.5 |
YLPM1
|
YLP motif containing 1 |
| chr10_-_128126405 | 0.05 |
ENST00000368654.8
|
MKI67
|
marker of proliferation Ki-67 |
| chr10_-_128126204 | 0.05 |
ENST00000368653.7
|
MKI67
|
marker of proliferation Ki-67 |
| chr1_-_39901996 | 0.05 |
ENST00000397332.2
|
MYCL
|
MYCL proto-oncogene, bHLH transcription factor |
| chr1_-_39901861 | 0.05 |
ENST00000372816.3
ENST00000372815.1 |
MYCL
|
MYCL proto-oncogene, bHLH transcription factor |
| chr1_-_197146620 | 0.05 |
ENST00000367409.9
ENST00000680265.1 |
ASPM
|
assembly factor for spindle microtubules |
| chr1_-_197146688 | 0.05 |
ENST00000294732.11
|
ASPM
|
assembly factor for spindle microtubules |
| chr9_-_137302264 | 0.05 |
ENST00000356628.4
|
NRARP
|
NOTCH regulated ankyrin repeat protein |
| chrX_-_130903224 | 0.05 |
ENST00000370935.5
|
ENOX2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr9_-_71768386 | 0.05 |
ENST00000377066.9
ENST00000377044.9 |
CEMIP2
|
cell migration inducing hyaluronidase 2 |
| chr4_+_112231748 | 0.05 |
ENST00000274000.10
ENST00000309703.10 |
AP1AR
|
adaptor related protein complex 1 associated regulatory protein |
| chr9_+_131125578 | 0.05 |
ENST00000359428.10
ENST00000411637.6 ENST00000652454.1 |
NUP214
|
nucleoporin 214 |
| chr22_+_49853801 | 0.05 |
ENST00000216268.6
|
ZBED4
|
zinc finger BED-type containing 4 |
| chr7_+_155298561 | 0.05 |
ENST00000476756.1
|
INSIG1
|
insulin induced gene 1 |
| chr8_-_20303955 | 0.05 |
ENST00000381569.5
|
LZTS1
|
leucine zipper tumor suppressor 1 |
| chr1_+_248038172 | 0.05 |
ENST00000366479.4
|
OR2L2
|
olfactory receptor family 2 subfamily L member 2 |
| chr11_+_60924452 | 0.05 |
ENST00000453848.7
ENST00000544065.5 ENST00000005286.8 |
TMEM132A
|
transmembrane protein 132A |
| chr19_+_7595830 | 0.05 |
ENST00000160298.9
ENST00000446248.4 |
CAMSAP3
|
calmodulin regulated spectrin associated protein family member 3 |
| chr9_-_137070548 | 0.05 |
ENST00000409687.5
|
SAPCD2
|
suppressor APC domain containing 2 |
| chr16_+_88570387 | 0.05 |
ENST00000452588.6
ENST00000301011.10 |
ZC3H18
|
zinc finger CCCH-type containing 18 |
| chr3_+_189789672 | 0.05 |
ENST00000434928.5
|
TP63
|
tumor protein p63 |
| chr1_-_116667668 | 0.05 |
ENST00000369486.8
ENST00000369483.5 |
IGSF3
|
immunoglobulin superfamily member 3 |
| chr6_-_105403062 | 0.05 |
ENST00000652536.2
|
PREP
|
prolyl endopeptidase |
| chr22_+_23180365 | 0.05 |
ENST00000359540.7
ENST00000305877.13 |
BCR
|
BCR activator of RhoGEF and GTPase |
| chr3_-_186109067 | 0.05 |
ENST00000306376.10
|
ETV5
|
ETS variant transcription factor 5 |
| chr3_+_124033356 | 0.05 |
ENST00000682506.1
|
KALRN
|
kalirin RhoGEF kinase |
| chr6_-_85643778 | 0.05 |
ENST00000676637.1
ENST00000678528.1 |
SYNCRIP
|
synaptotagmin binding cytoplasmic RNA interacting protein |
| chr9_+_27109135 | 0.05 |
ENST00000519097.5
ENST00000615002.4 |
TEK
|
TEK receptor tyrosine kinase |
| chr10_-_60572599 | 0.05 |
ENST00000503366.5
|
ANK3
|
ankyrin 3 |
| chr11_+_64186163 | 0.05 |
ENST00000305218.9
ENST00000538945.5 |
STIP1
|
stress induced phosphoprotein 1 |
| chr6_-_75206044 | 0.05 |
ENST00000322507.13
|
COL12A1
|
collagen type XII alpha 1 chain |
| chr11_-_44950151 | 0.05 |
ENST00000533940.5
ENST00000533937.1 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr9_+_4792971 | 0.05 |
ENST00000381732.3
ENST00000442869.5 |
RCL1
|
RNA terminal phosphate cyclase like 1 |
| chr9_+_105700953 | 0.05 |
ENST00000374688.5
|
TMEM38B
|
transmembrane protein 38B |
| chr11_+_67391975 | 0.05 |
ENST00000307980.7
|
RAD9A
|
RAD9 checkpoint clamp component A |
| chr18_-_13915531 | 0.05 |
ENST00000327606.4
|
MC2R
|
melanocortin 2 receptor |
| chrX_-_38220824 | 0.05 |
ENST00000378533.4
ENST00000432886.6 ENST00000544439.5 ENST00000538295.5 |
SRPX
|
sushi repeat containing protein X-linked |
| chr9_+_72577939 | 0.05 |
ENST00000645773.1
|
TMC1
|
transmembrane channel like 1 |
| chr10_+_123135938 | 0.05 |
ENST00000357878.7
|
HMX3
|
H6 family homeobox 3 |
| chr19_-_17264718 | 0.05 |
ENST00000431146.6
ENST00000594190.5 |
USHBP1
|
USH1 protein network component harmonin binding protein 1 |
| chr12_+_69585666 | 0.05 |
ENST00000543146.2
|
CCT2
|
chaperonin containing TCP1 subunit 2 |
| chr1_-_169485931 | 0.05 |
ENST00000367804.4
ENST00000646596.1 ENST00000236137.10 |
SLC19A2
|
solute carrier family 19 member 2 |
| chr7_+_5592805 | 0.05 |
ENST00000382361.8
|
FSCN1
|
fascin actin-bundling protein 1 |
| chr7_-_140176970 | 0.05 |
ENST00000397560.7
|
KDM7A
|
lysine demethylase 7A |
| chr17_+_7857695 | 0.05 |
ENST00000571846.5
|
CYB5D1
|
cytochrome b5 domain containing 1 |
| chr11_-_119729158 | 0.05 |
ENST00000264025.8
|
NECTIN1
|
nectin cell adhesion molecule 1 |
| chrM_+_4467 | 0.05 |
ENST00000361453.3
|
MT-ND2
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 2 |
| chr7_+_2631978 | 0.05 |
ENST00000258796.12
|
TTYH3
|
tweety family member 3 |
| chr12_-_23951020 | 0.05 |
ENST00000441133.2
ENST00000545921.5 |
SOX5
|
SRY-box transcription factor 5 |
| chr8_-_100953331 | 0.05 |
ENST00000353245.7
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta |
| chr6_-_112087451 | 0.05 |
ENST00000368662.10
|
TUBE1
|
tubulin epsilon 1 |
| chr7_-_151227035 | 0.05 |
ENST00000441774.1
ENST00000287844.7 ENST00000222388.6 |
ABCF2
ABCF2-H2BE1
|
ATP binding cassette subfamily F member 2 ABCF2-H2BE1 readthrough |
| chr1_+_200739542 | 0.05 |
ENST00000358823.6
|
CAMSAP2
|
calmodulin regulated spectrin associated protein family member 2 |
| chr4_-_82429402 | 0.05 |
ENST00000602300.5
|
HNRNPDL
|
heterogeneous nuclear ribonucleoprotein D like |
| chr6_-_53348902 | 0.04 |
ENST00000370913.5
ENST00000304434.11 |
ELOVL5
|
ELOVL fatty acid elongase 5 |
| chr16_-_48247533 | 0.04 |
ENST00000356608.7
ENST00000569991.1 |
ABCC11
|
ATP binding cassette subfamily C member 11 |
| chr9_+_131125973 | 0.04 |
ENST00000651639.1
ENST00000451030.5 ENST00000531584.1 |
NUP214
|
nucleoporin 214 |
| chr8_-_100952918 | 0.04 |
ENST00000395957.6
ENST00000395948.6 ENST00000457309.2 |
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta |
| chr7_+_116672187 | 0.04 |
ENST00000318493.11
ENST00000397752.8 |
MET
|
MET proto-oncogene, receptor tyrosine kinase |
| chr16_-_27549887 | 0.04 |
ENST00000561623.5
ENST00000356183.9 |
GTF3C1
|
general transcription factor IIIC subunit 1 |
| chr14_-_21048431 | 0.04 |
ENST00000555026.5
|
NDRG2
|
NDRG family member 2 |
| chr7_-_149028651 | 0.04 |
ENST00000286091.9
|
PDIA4
|
protein disulfide isomerase family A member 4 |
| chr5_-_43313403 | 0.04 |
ENST00000325110.11
|
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 |
| chr5_+_51383394 | 0.04 |
ENST00000230658.12
|
ISL1
|
ISL LIM homeobox 1 |
| chr7_-_16881967 | 0.04 |
ENST00000402239.7
ENST00000310398.7 ENST00000414935.1 |
AGR3
|
anterior gradient 3, protein disulphide isomerase family member |
| chrX_+_83508284 | 0.04 |
ENST00000644024.2
|
POU3F4
|
POU class 3 homeobox 4 |
| chr17_-_51046868 | 0.04 |
ENST00000510283.5
ENST00000510855.1 |
SPAG9
|
sperm associated antigen 9 |
| chr11_+_64186219 | 0.04 |
ENST00000543847.1
|
STIP1
|
stress induced phosphoprotein 1 |
| chr7_+_107583919 | 0.04 |
ENST00000491150.5
|
BCAP29
|
B cell receptor associated protein 29 |
| chr11_+_119085200 | 0.04 |
ENST00000650101.1
|
HMBS
|
hydroxymethylbilane synthase |
| chr2_-_55050556 | 0.04 |
ENST00000394611.6
|
RTN4
|
reticulon 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX9
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0032600 | B cell receptor transport within lipid bilayer(GO:0032595) B cell receptor transport into membrane raft(GO:0032597) protein transport out of membrane raft(GO:0032599) chemokine receptor transport out of membrane raft(GO:0032600) negative regulation of transforming growth factor beta3 production(GO:0032913) chemokine receptor transport within lipid bilayer(GO:0033606) |
| 0.1 | 0.3 | GO:0008614 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 biosynthetic process(GO:0042819) |
| 0.1 | 0.3 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 0.2 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.1 | 0.2 | GO:0051710 | cytolysis by symbiont of host cells(GO:0001897) regulation of cytolysis in other organism(GO:0051710) |
| 0.1 | 0.5 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.2 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.1 | GO:0032912 | negative regulation of transforming growth factor beta2 production(GO:0032912) |
| 0.0 | 0.2 | GO:0009099 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 | 0.6 | GO:0070444 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.1 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.0 | 0.3 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.2 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.1 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.0 | 0.3 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.0 | 0.1 | GO:0016476 | calcium-dependent cell-matrix adhesion(GO:0016340) regulation of embryonic cell shape(GO:0016476) |
| 0.0 | 0.1 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.2 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:1903490 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.0 | GO:0097272 | ammonia homeostasis(GO:0097272) urea homeostasis(GO:0097274) |
| 0.0 | 0.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.1 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.0 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.0 | 0.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.0 | 0.0 | GO:1901258 | cardiac cell fate determination(GO:0060913) positive regulation of granulocyte colony-stimulating factor production(GO:0071657) positive regulation of macrophage colony-stimulating factor production(GO:1901258) |
| 0.0 | 0.1 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.1 | GO:0033490 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.0 | 0.0 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.0 | 0.0 | GO:0098583 | mastication(GO:0071626) learned vocalization behavior(GO:0098583) |
| 0.0 | 0.1 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.1 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.0 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.0 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.0 | 0.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.2 | GO:2000586 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) |
| 0.0 | 0.0 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.0 | GO:1900222 | negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.0 | GO:1903004 | flavin adenine dinucleotide metabolic process(GO:0072387) regulation of protein K63-linked deubiquitination(GO:1903004) positive regulation of protein K63-linked deubiquitination(GO:1903006) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.1 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.1 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.2 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.3 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.5 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.1 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.0 | 0.4 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.0 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.0 | GO:0044393 | microspike(GO:0044393) |
| 0.0 | 0.7 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.0 | 0.2 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.2 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.1 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.0 | 0.1 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) cholestenol delta-isomerase activity(GO:0047750) |
| 0.0 | 0.1 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.3 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.1 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.0 | 0.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.1 | GO:0004420 | hydroxymethylglutaryl-CoA reductase (NADPH) activity(GO:0004420) hydroxymethylglutaryl-CoA reductase activity(GO:0042282) |
| 0.0 | 0.1 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.7 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.0 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.0 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.1 | GO:0043273 | CTPase activity(GO:0043273) |
| 0.0 | 0.0 | GO:0000992 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.0 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.0 | 0.5 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |