chr10_-_104085847
Show fit
0.56
ENST00000648076.2
COL17A1
collagen type XVII alpha 1 chain
chr3_+_189789643
Show fit
0.45
ENST00000354600.10
TP63
tumor protein p63
chr12_-_8662619
Show fit
0.40
ENST00000544889.1
ENST00000543369.5
MFAP5
microfibril associated protein 5
chr12_-_8662703
Show fit
0.38
ENST00000535336.5
MFAP5
microfibril associated protein 5
chr6_-_106975452
Show fit
0.34
ENST00000619869.1
ENST00000619133.4
CD24
CD24 molecule
chr6_-_106975309
Show fit
0.33
ENST00000615659.1
CD24
CD24 molecule
chr12_-_8662808
Show fit
0.31
ENST00000359478.7
ENST00000396549.6
MFAP5
microfibril associated protein 5
chr8_+_94641074
Show fit
0.30
ENST00000423620.6
ESRP1
epithelial splicing regulatory protein 1
chr12_-_27971970
Show fit
0.29
ENST00000395872.5
ENST00000201015.8
PTHLH
parathyroid hormone like hormone
chr13_+_112979306
Show fit
0.27
ENST00000421756.5
MCF2L
MCF.2 cell line derived transforming sequence like
chr1_-_177164673
Show fit
0.23
ENST00000424564.2
ENST00000361833.7
ASTN1
astrotactin 1
chr3_-_123620496
Show fit
0.22
ENST00000578202.1
MYLK
myosin light chain kinase
chr3_-_123620571
Show fit
0.22
ENST00000583087.5
MYLK
myosin light chain kinase
chr3_+_189789734
Show fit
0.22
ENST00000437221.5
ENST00000392463.6
ENST00000392461.7
ENST00000449992.5
ENST00000456148.1
TP63
tumor protein p63
chr4_-_110198650
Show fit
0.21
ENST00000394607.7
ELOVL6
ELOVL fatty acid elongase 6
chr4_-_110198579
Show fit
0.21
ENST00000302274.8
ELOVL6
ELOVL fatty acid elongase 6
chr5_+_140868945
Show fit
0.21
ENST00000398640.7
PCDHA11
protocadherin alpha 11
chr12_-_8662881
Show fit
0.21
ENST00000433590.6
MFAP5
microfibril associated protein 5
chr12_-_95217373
Show fit
0.19
ENST00000549499.1
ENST00000546711.5
ENST00000343958.9
FGD6
FYVE, RhoGEF and PH domain containing 6
chr17_+_9021501
Show fit
0.18
ENST00000173229.7
NTN1
netrin 1
chr9_-_23826231
Show fit
0.17
ENST00000397312.7
ELAVL2
ELAV like RNA binding protein 2
chr7_-_93226449
Show fit
0.17
ENST00000394468.7
ENST00000453812.2
HEPACAM2
HEPACAM family member 2
chr8_-_42501224
Show fit
0.17
ENST00000520262.6
ENST00000517366.1
SLC20A2
solute carrier family 20 member 2
chr10_-_14572123
Show fit
0.17
ENST00000378465.7
ENST00000452706.6
ENST00000622567.4
ENST00000378458.6
FAM107B
family with sequence similarity 107 member B
chr11_-_115504389
Show fit
0.16
ENST00000545380.5
ENST00000452722.7
ENST00000331581.11
ENST00000537058.5
ENST00000536727.5
ENST00000542447.6
CADM1
cell adhesion molecule 1
chr15_-_59372863
Show fit
0.16
ENST00000288235.9
MYO1E
myosin IE
chr5_-_24644968
Show fit
0.16
ENST00000264463.8
CDH10
cadherin 10
chr9_-_71768386
Show fit
0.16
ENST00000377066.9
ENST00000377044.9
CEMIP2
cell migration inducing hyaluronidase 2
chr3_+_39467598
Show fit
0.15
ENST00000428261.5
ENST00000420739.5
ENST00000415443.5
ENST00000447324.5
ENST00000383754.7
MOBP
myelin associated oligodendrocyte basic protein
chr8_+_66493556
Show fit
0.15
ENST00000305454.8
ENST00000522977.5
ENST00000480005.1
VXN
vexin
chr3_+_39467672
Show fit
0.15
ENST00000436143.6
ENST00000441980.6
ENST00000682069.1
ENST00000311042.10
MOBP
myelin associated oligodendrocyte basic protein
chr12_-_85836372
Show fit
0.15
ENST00000361228.5
RASSF9
Ras association domain family member 9
chr12_-_91058016
Show fit
0.14
ENST00000266719.4
KERA
keratocan
chr6_+_156777366
Show fit
0.14
ENST00000636930.2
ARID1B
AT-rich interaction domain 1B
chr1_+_84164684
Show fit
0.14
ENST00000370680.5
PRKACB
protein kinase cAMP-activated catalytic subunit beta
chr2_+_195657179
Show fit
0.13
ENST00000359634.10
SLC39A10
solute carrier family 39 member 10
chr3_+_189789672
Show fit
0.13
ENST00000434928.5
TP63
tumor protein p63
chr4_-_102345469
Show fit
0.13
ENST00000356736.5
ENST00000682932.1
SLC39A8
solute carrier family 39 member 8
chr8_-_133297092
Show fit
0.13
ENST00000522890.5
ENST00000675983.1
ENST00000518176.5
ENST00000323851.13
ENST00000522476.5
ENST00000518066.5
ENST00000521544.5
ENST00000674605.1
ENST00000518480.5
ENST00000523892.5
NDRG1
N-myc downstream regulated 1
chr11_-_119729158
Show fit
0.13
ENST00000264025.8
NECTIN1
nectin cell adhesion molecule 1
chr4_+_15427998
Show fit
0.12
ENST00000444304.3
C1QTNF7
C1q and TNF related 7
chr17_-_51046868
Show fit
0.12
ENST00000510283.5
ENST00000510855.1
SPAG9
sperm associated antigen 9
chr4_+_39406917
Show fit
0.12
ENST00000257408.5
KLB
klotho beta
chr1_-_217137748
Show fit
0.12
ENST00000493603.5
ENST00000366940.6
ESRRG
estrogen related receptor gamma
chr10_+_113553039
Show fit
0.12
ENST00000351270.4
HABP2
hyaluronan binding protein 2
chr12_-_122500520
Show fit
0.11
ENST00000540586.1
ENST00000543897.5
ZCCHC8
zinc finger CCHC-type containing 8
chr2_-_207165923
Show fit
0.11
ENST00000309446.11
KLF7
Kruppel like factor 7
chr17_-_39607876
Show fit
0.11
ENST00000302584.5
NEUROD2
neuronal differentiation 2
chr12_-_122500924
Show fit
0.11
ENST00000633063.3
ZCCHC8
zinc finger CCHC-type containing 8
chr11_+_57667747
Show fit
0.11
ENST00000527985.5
ZDHHC5
zinc finger DHHC-type palmitoyltransferase 5
chr11_+_131911396
Show fit
0.11
ENST00000425719.6
ENST00000374784.5
NTM
neurotrimin
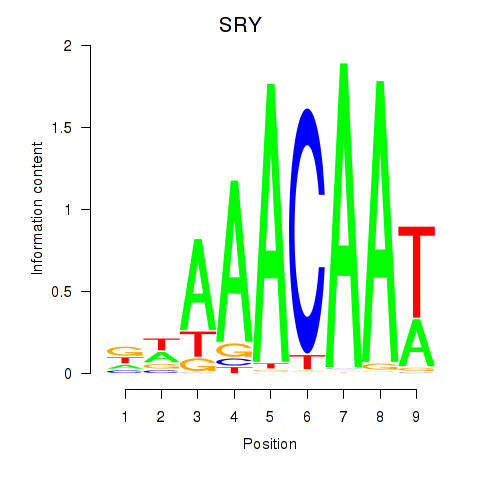
chr11_-_16356538
Show fit
0.11
ENST00000683767.1
SOX6
SRY-box transcription factor 6
chr3_-_192917832
Show fit
0.11
ENST00000392452.3
MB21D2
Mab-21 domain containing 2
chr12_-_122500947
Show fit
0.11
ENST00000672018.1
ZCCHC8
zinc finger CCHC-type containing 8
chr6_-_42451261
Show fit
0.11
ENST00000372917.8
ENST00000340840.6
ENST00000354325.2
TRERF1
transcriptional regulating factor 1
chr13_-_46142834
Show fit
0.11
ENST00000674665.1
LCP1
lymphocyte cytosolic protein 1
chr12_-_10098977
Show fit
0.11
ENST00000315330.8
ENST00000457018.6
CLEC1A
C-type lectin domain family 1 member A
chr10_-_60572599
Show fit
0.10
ENST00000503366.5
ANK3
ankyrin 3
chrX_+_78747705
Show fit
0.10
ENST00000614823.5
ENST00000435339.3
ENST00000514744.5
LPAR4
lysophosphatidic acid receptor 4
chr3_-_33659097
Show fit
0.10
ENST00000461133.8
ENST00000496954.2
CLASP2
cytoplasmic linker associated protein 2
chr9_-_122227525
Show fit
0.10
ENST00000373755.6
ENST00000373754.6
LHX6
LIM homeobox 6
chr2_+_27442365
Show fit
0.10
ENST00000543753.5
ENST00000288873.7
KRTCAP3
keratinocyte associated protein 3
chr2_+_27442421
Show fit
0.10
ENST00000407293.5
KRTCAP3
keratinocyte associated protein 3
chrX_-_24672654
Show fit
0.10
ENST00000379145.5
PCYT1B
phosphate cytidylyltransferase 1, choline, beta
chr1_-_39901861
Show fit
0.10
ENST00000372816.3
ENST00000372815.1
MYCL
MYCL proto-oncogene, bHLH transcription factor
chr9_-_127847117
Show fit
0.10
ENST00000480266.5
ENG
endoglin
chr9_+_121699328
Show fit
0.10
ENST00000373782.7
DAB2IP
DAB2 interacting protein
chr18_-_55635948
Show fit
0.10
ENST00000565124.4
ENST00000398339.5
TCF4
transcription factor 4
chr1_-_217076889
Show fit
0.10
ENST00000493748.5
ENST00000463665.5
ESRRG
estrogen related receptor gamma
chr9_-_39239174
Show fit
0.10
ENST00000358144.6
CNTNAP3
contactin associated protein family member 3
chr1_-_116667668
Show fit
0.10
ENST00000369486.8
ENST00000369483.5
IGSF3
immunoglobulin superfamily member 3
chr3_+_142623386
Show fit
0.09
ENST00000337777.7
ENST00000497199.5
PLS1
plastin 1
chr15_-_34318761
Show fit
0.09
ENST00000290209.9
SLC12A6
solute carrier family 12 member 6
chrX_-_111412162
Show fit
0.09
ENST00000637570.1
ENST00000356220.8
ENST00000636035.2
ENST00000637453.1
ENST00000635795.1
DCX
doublecortin
chr9_-_23826299
Show fit
0.09
ENST00000380117.5
ELAVL2
ELAV like RNA binding protein 2
chr3_-_33659441
Show fit
0.09
ENST00000650653.1
ENST00000480013.6
CLASP2
cytoplasmic linker associated protein 2
chr18_-_55321986
Show fit
0.09
ENST00000570287.6
TCF4
transcription factor 4
chr1_-_93180261
Show fit
0.09
ENST00000370280.1
ENST00000479918.5
TMED5
transmembrane p24 trafficking protein 5
chr1_+_147902789
Show fit
0.09
ENST00000369235.2
GJA8
gap junction protein alpha 8
chr6_-_116060859
Show fit
0.09
ENST00000606080.2
FRK
fyn related Src family tyrosine kinase
chr4_-_173530219
Show fit
0.09
ENST00000359562.4
HAND2
Links
heart and neural crest derivatives expressed 2
chr8_+_24294107
Show fit
0.08
ENST00000437154.6
ADAM28
ADAM metallopeptidase domain 28
chr19_+_10602436
Show fit
0.08
ENST00000590382.5
ENST00000407327.8
SLC44A2
solute carrier family 44 member 2
chr14_-_22819721
Show fit
0.08
ENST00000554517.5
ENST00000285850.11
ENST00000397529.6
ENST00000555702.5
SLC7A7
solute carrier family 7 member 7
chr8_-_81483226
Show fit
0.08
ENST00000256104.5
FABP4
fatty acid binding protein 4
chr18_-_13915531
Show fit
0.08
ENST00000327606.4
MC2R
melanocortin 2 receptor
chr20_-_22585451
Show fit
0.08
ENST00000377115.4
FOXA2
forkhead box A2
chr5_-_138338325
Show fit
0.08
ENST00000510119.1
ENST00000513970.5
CDC25C
cell division cycle 25C
chr1_-_39901996
Show fit
0.08
ENST00000397332.2
MYCL
MYCL proto-oncogene, bHLH transcription factor
chrX_+_108044967
Show fit
0.08
ENST00000415430.7
VSIG1
V-set and immunoglobulin domain containing 1
chr14_+_22281097
Show fit
0.08
ENST00000390465.2
TRAV38-2DV8
T cell receptor alpha variable 38-2/delta variable 8
chr12_-_31324129
Show fit
0.08
ENST00000454658.6
SINHCAF
SIN3-HDAC complex associated factor
chr18_+_58045642
Show fit
0.08
ENST00000676223.1
ENST00000675147.1
NEDD4L
NEDD4 like E3 ubiquitin protein ligase
chr12_-_24562438
Show fit
0.07
ENST00000646273.1
ENST00000659413.1
ENST00000446891.7
SOX5
SRY-box transcription factor 5
chr6_-_52244500
Show fit
0.07
ENST00000336123.5
IL17F
interleukin 17F
chr7_-_100100716
Show fit
0.07
ENST00000354230.7
ENST00000425308.5
MCM7
minichromosome maintenance complex component 7
chr16_-_68448491
Show fit
0.07
ENST00000561749.1
ENST00000219334.10
SMPD3
sphingomyelin phosphodiesterase 3
chr2_-_71227055
Show fit
0.07
ENST00000244221.9
PAIP2B
poly(A) binding protein interacting protein 2B
chr10_-_60389833
Show fit
0.07
ENST00000280772.7
ANK3
ankyrin 3
chrX_+_108045050
Show fit
0.07
ENST00000458383.1
ENST00000217957.10
VSIG1
V-set and immunoglobulin domain containing 1
chr3_+_57756230
Show fit
0.07
ENST00000295951.7
ENST00000659705.1
ENST00000671191.1
SLMAP
sarcolemma associated protein
chr15_-_53733103
Show fit
0.07
ENST00000559418.5
WDR72
WD repeat domain 72
chr19_+_13023958
Show fit
0.07
ENST00000587760.5
ENST00000585575.5
NFIX
nuclear factor I X
chr1_+_93179883
Show fit
0.07
ENST00000343253.11
CCDC18
coiled-coil domain containing 18
chr6_+_50713526
Show fit
0.07
ENST00000008391.4
TFAP2D
transcription factor AP-2 delta
chr2_-_55010348
Show fit
0.07
ENST00000394609.6
RTN4
reticulon 4
chrM_-_14669
Show fit
0.07
ENST00000361681.2
MT-ND6
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 6
chr7_-_149028651
Show fit
0.07
ENST00000286091.9
PDIA4
protein disulfide isomerase family A member 4
chr12_+_95217792
Show fit
0.07
ENST00000436874.6
ENST00000551472.5
ENST00000552821.5
VEZT
vezatin, adherens junctions transmembrane protein
chr10_+_11164961
Show fit
0.07
ENST00000399850.7
ENST00000417956.6
CELF2
CUGBP Elav-like family member 2
chr18_+_58045683
Show fit
0.07
ENST00000592846.5
ENST00000675801.1
NEDD4L
NEDD4 like E3 ubiquitin protein ligase
chr1_+_206834347
Show fit
0.07
ENST00000340758.7
IL19
interleukin 19
chr4_+_70592295
Show fit
0.07
ENST00000449493.2
AMBN
ameloblastin
chr11_+_112961247
Show fit
0.07
ENST00000621518.4
ENST00000618266.4
ENST00000615112.4
ENST00000615285.4
ENST00000619839.4
ENST00000401611.6
ENST00000621128.4
NCAM1
neural cell adhesion molecule 1
chr1_+_99850485
Show fit
0.07
ENST00000370165.7
ENST00000370163.7
ENST00000294724.8
AGL
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase
chr7_-_149028452
Show fit
0.06
ENST00000413966.1
ENST00000652332.1
PDIA4
protein disulfide isomerase family A member 4
chr1_+_159439722
Show fit
0.06
ENST00000641630.1
ENST00000423932.6
OR10J1
olfactory receptor family 10 subfamily J member 1
chr9_+_27109135
Show fit
0.06
ENST00000519097.5
ENST00000615002.4
TEK
TEK receptor tyrosine kinase
chr12_+_389334
Show fit
0.06
ENST00000540180.5
ENST00000422000.5
ENST00000535052.5
CCDC77
coiled-coil domain containing 77
chr10_-_60141004
Show fit
0.06
ENST00000355288.6
ANK3
ankyrin 3
chr9_+_74497308
Show fit
0.06
ENST00000376896.8
RORB
RAR related orphan receptor B
chr9_+_130693756
Show fit
0.06
ENST00000546165.5
ENST00000372352.7
ENST00000372358.10
ENST00000372351.7
ENST00000372350.7
ENST00000495699.2
EXOSC2
exosome component 2
chr15_+_90001300
Show fit
0.06
ENST00000268154.9
ZNF710
zinc finger protein 710
chr2_-_231125032
Show fit
0.06
ENST00000258400.4
HTR2B
5-hydroxytryptamine receptor 2B
chr8_-_86743626
Show fit
0.06
ENST00000320005.6
CNGB3
cyclic nucleotide gated channel subunit beta 3
chr12_-_102478539
Show fit
0.06
ENST00000424202.6
IGF1
insulin like growth factor 1
chr8_+_24294044
Show fit
0.06
ENST00000265769.9
ADAM28
ADAM metallopeptidase domain 28
chrX_-_19670983
Show fit
0.06
ENST00000379716.5
SH3KBP1
SH3 domain containing kinase binding protein 1
chr11_-_30586866
Show fit
0.06
ENST00000528686.2
MPPED2
metallophosphoesterase domain containing 2
chr11_+_112961480
Show fit
0.06
ENST00000621850.4
NCAM1
neural cell adhesion molecule 1
chr5_+_87267792
Show fit
0.06
ENST00000274376.11
RASA1
RAS p21 protein activator 1
chr12_+_59664677
Show fit
0.06
ENST00000548610.5
SLC16A7
solute carrier family 16 member 7
chr2_+_190408324
Show fit
0.06
ENST00000417958.5
ENST00000432036.5
ENST00000392328.6
MFSD6
major facilitator superfamily domain containing 6
chr9_+_27109393
Show fit
0.06
ENST00000406359.8
TEK
TEK receptor tyrosine kinase
chr4_+_164754116
Show fit
0.06
ENST00000507311.1
SMIM31
small integral membrane protein 31
chr18_+_33578213
Show fit
0.06
ENST00000681521.1
ENST00000269197.12
ASXL3
ASXL transcriptional regulator 3
chr3_+_113211459
Show fit
0.06
ENST00000495514.5
BOC
BOC cell adhesion associated, oncogene regulated
chr18_-_55589795
Show fit
0.05
ENST00000568740.5
ENST00000629387.2
TCF4
transcription factor 4
chr7_+_143183419
Show fit
0.05
ENST00000446620.1
TAS2R39
taste 2 receptor member 39
chr11_-_44950151
Show fit
0.05
ENST00000533940.5
ENST00000533937.1
TP53I11
tumor protein p53 inducible protein 11
chr12_-_102480638
Show fit
0.05
ENST00000392904.5
IGF1
insulin like growth factor 1
chr18_-_55322215
Show fit
0.05
ENST00000457482.7
TCF4
transcription factor 4
chr11_+_112961402
Show fit
0.05
ENST00000613217.4
ENST00000316851.12
ENST00000620046.4
ENST00000531044.5
ENST00000529356.5
NCAM1
neural cell adhesion molecule 1
chr2_+_69013414
Show fit
0.05
ENST00000681816.1
ENST00000482235.2
ANTXR1
ANTXR cell adhesion molecule 1
chr22_+_39901075
Show fit
0.05
ENST00000344138.9
GRAP2
GRB2 related adaptor protein 2
chr6_+_156777882
Show fit
0.05
ENST00000350026.10
ENST00000647938.1
ENST00000674298.1
ARID1B
AT-rich interaction domain 1B
chr18_-_55589836
Show fit
0.05
ENST00000537578.5
ENST00000564403.6
TCF4
transcription factor 4
chr20_-_44521989
Show fit
0.05
ENST00000342374.5
ENST00000255175.5
SERINC3
serine incorporator 3
chr15_+_89776326
Show fit
0.05
ENST00000341735.5
MESP2
mesoderm posterior bHLH transcription factor 2
chr17_-_17972374
Show fit
0.05
ENST00000318094.14
ENST00000540946.5
ENST00000379504.8
ENST00000542206.5
ENST00000395739.8
ENST00000581396.5
ENST00000535933.5
ENST00000579586.1
TOM1L2
target of myb1 like 2 membrane trafficking protein
chr12_-_48570046
Show fit
0.05
ENST00000301046.6
ENST00000549817.1
LALBA
lactalbumin alpha
chr11_+_128693887
Show fit
0.05
ENST00000281428.12
FLI1
Fli-1 proto-oncogene, ETS transcription factor
chr16_-_4273014
Show fit
0.05
ENST00000204517.11
TFAP4
transcription factor AP-4
chr17_-_81239085
Show fit
0.05
ENST00000300714.7
TEPSIN
TEPSIN adaptor related protein complex 4 accessory protein
chr6_-_162727748
Show fit
0.05
ENST00000366892.5
ENST00000366898.6
ENST00000366897.5
ENST00000366896.5
ENST00000674250.1
PRKN
parkin RBR E3 ubiquitin protein ligase
chr14_+_58427970
Show fit
0.05
ENST00000261244.9
KIAA0586
KIAA0586
chr5_-_11589019
Show fit
0.05
ENST00000511377.5
CTNND2
catenin delta 2
chr4_-_64409444
Show fit
0.05
ENST00000381210.8
ENST00000507440.5
TECRL
trans-2,3-enoyl-CoA reductase like
chr12_-_81369348
Show fit
0.05
ENST00000548670.1
ENST00000541017.5
ENST00000541570.6
ENST00000553058.5
PPFIA2
PTPRF interacting protein alpha 2
chr5_-_147782681
Show fit
0.05
ENST00000616793.5
ENST00000333010.6
ENST00000265272.9
JAKMIP2
janus kinase and microtubule interacting protein 2
chr18_-_55302613
Show fit
0.05
ENST00000561831.7
TCF4
transcription factor 4
chr11_+_128694052
Show fit
0.05
ENST00000527786.7
ENST00000534087.3
FLI1
Fli-1 proto-oncogene, ETS transcription factor
chr9_+_27109200
Show fit
0.04
ENST00000380036.10
TEK
TEK receptor tyrosine kinase
chr10_-_50279715
Show fit
0.04
ENST00000395526.9
ASAH2
N-acylsphingosine amidohydrolase 2
chr17_+_38705482
Show fit
0.04
ENST00000620609.4
MLLT6
MLLT6, PHD finger containing
chrX_+_123961696
Show fit
0.04
ENST00000371145.8
ENST00000371157.7
ENST00000371144.7
STAG2
stromal antigen 2
chr4_-_112516176
Show fit
0.04
ENST00000313341.4
NEUROG2
neurogenin 2
chr15_+_59105099
Show fit
0.04
ENST00000288207.7
ENST00000559622.5
CCNB2
cyclin B2
chrM_+_8489
Show fit
0.04
ENST00000361899.2
MT-ATP6
mitochondrially encoded ATP synthase membrane subunit 6
chr1_+_25338294
Show fit
0.04
ENST00000374358.5
TMEM50A
transmembrane protein 50A
chr2_-_159616442
Show fit
0.04
ENST00000541068.6
ENST00000392783.7
ENST00000392782.5
BAZ2B
bromodomain adjacent to zinc finger domain 2B
chr17_-_81239025
Show fit
0.04
ENST00000637944.2
TEPSIN
TEPSIN adaptor related protein complex 4 accessory protein
chr12_-_102480552
Show fit
0.04
ENST00000337514.11
ENST00000307046.8
IGF1
insulin like growth factor 1
chr3_+_178536205
Show fit
0.04
ENST00000420517.6
KCNMB2
potassium calcium-activated channel subfamily M regulatory beta subunit 2
chr2_+_9961165
Show fit
0.04
ENST00000405379.6
GRHL1
grainyhead like transcription factor 1
chr4_+_109827963
Show fit
0.04
ENST00000317735.7
RRH
retinal pigment epithelium-derived rhodopsin homolog
chr14_+_58427686
Show fit
0.04
ENST00000650904.1
ENST00000652326.2
ENST00000554463.5
ENST00000555833.5
KIAA0586
KIAA0586
chr18_+_58341038
Show fit
0.04
ENST00000679791.1
NEDD4L
NEDD4 like E3 ubiquitin protein ligase
chr10_-_125160499
Show fit
0.04
ENST00000494626.6
ENST00000337195.9
CTBP2
C-terminal binding protein 2
chr3_+_41194741
Show fit
0.04
ENST00000643541.1
ENST00000426215.5
ENST00000645210.1
ENST00000646381.1
ENST00000405570.6
ENST00000642248.1
ENST00000433400.6
CTNNB1
catenin beta 1
chr18_+_21363593
Show fit
0.04
ENST00000580732.6
GREB1L
GREB1 like retinoic acid receptor coactivator
chr4_-_67883987
Show fit
0.04
ENST00000283916.11
TMPRSS11D
transmembrane serine protease 11D
chr3_+_124384513
Show fit
0.04
ENST00000682540.1
ENST00000522553.6
ENST00000682695.1
ENST00000682674.1
ENST00000684382.1
KALRN
kalirin RhoGEF kinase
chr12_+_55019967
Show fit
0.04
ENST00000242994.4
NEUROD4
neuronal differentiation 4
chrX_+_101488044
Show fit
0.04
ENST00000423738.4
ARMCX4
armadillo repeat containing X-linked 4
chr21_-_31558977
Show fit
0.04
ENST00000286827.7
ENST00000541036.5
TIAM1
TIAM Rac1 associated GEF 1
chr18_-_55589770
Show fit
0.04
ENST00000565018.6
ENST00000636400.2
TCF4
transcription factor 4
chr14_-_56805648
Show fit
0.04
ENST00000554788.5
ENST00000554845.1
ENST00000408990.8
OTX2
orthodenticle homeobox 2
chr2_+_86441341
Show fit
0.03
ENST00000312912.10
ENST00000409064.5
KDM3A
lysine demethylase 3A
chr11_-_111766359
Show fit
0.03
ENST00000341980.10
ENST00000311129.9
ENST00000393055.6
ENST00000426998.6
ENST00000527614.6
PPP2R1B
protein phosphatase 2 scaffold subunit Abeta
chrX_-_136880715
Show fit
0.03
ENST00000431446.7
ENST00000320676.11
ENST00000562646.5
RBMX
RNA binding motif protein X-linked
chr15_+_59105205
Show fit
0.03
ENST00000621385.1
CCNB2
cyclin B2
chr7_-_27180230
Show fit
0.03
ENST00000396344.4
HOXA10
homeobox A10
chr14_+_22516273
Show fit
0.03
ENST00000390510.1
TRAJ27
T cell receptor alpha joining 27
chr4_+_169660062
Show fit
0.03
ENST00000507875.5
ENST00000613795.4
CLCN3
chloride voltage-gated channel 3
chr1_+_165827786
Show fit
0.03
ENST00000642653.1
UCK2
uridine-cytidine kinase 2
chr5_-_131796965
Show fit
0.03
ENST00000514667.1
ENST00000511848.1
ENST00000510461.6
ENSG00000273217.1
FNIP1
novel protein
chr15_-_85794902
Show fit
0.03
ENST00000337975.6
KLHL25
kelch like family member 25
chr10_+_72893734
Show fit
0.03
ENST00000334011.10
OIT3
oncoprotein induced transcript 3
chr1_+_231162052
Show fit
0.03
ENST00000366653.6
ENST00000444294.7
TRIM67
tripartite motif containing 67