Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
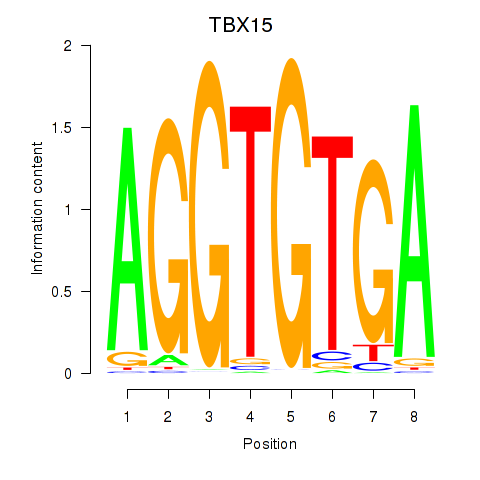
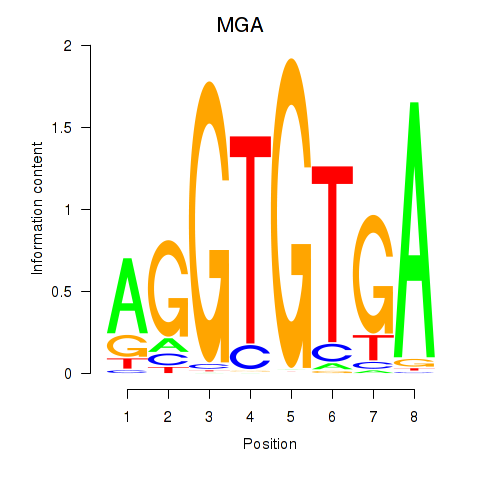
Results for TBX15_MGA
Z-value: 0.10
Transcription factors associated with TBX15_MGA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX15
|
ENSG00000092607.15 | T-box transcription factor 15 |
|
MGA
|
ENSG00000174197.17 | MAX dimerization protein MGA |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBX15 | hg38_v1_chr1_-_118988301_118988348, hg38_v1_chr1_-_118989504_118989556 | -0.78 | 2.3e-02 | Click! |
| MGA | hg38_v1_chr15_+_41660397_41660471 | 0.15 | 7.3e-01 | Click! |
Activity profile of TBX15_MGA motif
Sorted Z-values of TBX15_MGA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_35031263 | 0.25 |
ENST00000640135.1
ENST00000596348.2 |
SCN1B
|
sodium voltage-gated channel beta subunit 1 |
| chr10_+_92691897 | 0.24 |
ENST00000492654.3
|
HHEX
|
hematopoietically expressed homeobox |
| chr20_-_22584547 | 0.23 |
ENST00000419308.7
|
FOXA2
|
forkhead box A2 |
| chr19_-_45914775 | 0.22 |
ENST00000341294.4
|
NANOS2
|
nanos C2HC-type zinc finger 2 |
| chr22_-_41946729 | 0.20 |
ENST00000402420.1
|
CENPM
|
centromere protein M |
| chr11_-_111879154 | 0.20 |
ENST00000260257.9
|
FDXACB1
|
ferredoxin-fold anticodon binding domain containing 1 |
| chr5_+_55024250 | 0.18 |
ENST00000231009.3
|
GZMK
|
granzyme K |
| chr12_-_116276759 | 0.17 |
ENST00000548743.2
|
MED13L
|
mediator complex subunit 13L |
| chr7_-_76626127 | 0.16 |
ENST00000454397.1
|
POMZP3
|
POM121 and ZP3 fusion |
| chr17_-_8163522 | 0.16 |
ENST00000404970.3
|
VAMP2
|
vesicle associated membrane protein 2 |
| chr1_+_26410809 | 0.15 |
ENST00000254231.4
ENST00000326279.11 |
LIN28A
|
lin-28 homolog A |
| chr19_-_6416773 | 0.15 |
ENST00000595223.5
|
KHSRP
|
KH-type splicing regulatory protein |
| chr4_-_65670339 | 0.15 |
ENST00000273854.7
|
EPHA5
|
EPH receptor A5 |
| chr4_-_65670478 | 0.15 |
ENST00000613740.5
ENST00000622150.4 ENST00000511294.1 |
EPHA5
|
EPH receptor A5 |
| chr7_-_44140372 | 0.14 |
ENST00000446581.5
|
MYL7
|
myosin light chain 7 |
| chr12_+_18738102 | 0.14 |
ENST00000317658.5
|
CAPZA3
|
capping actin protein of muscle Z-line subunit alpha 3 |
| chr2_+_202073282 | 0.14 |
ENST00000459709.5
|
KIAA2012
|
KIAA2012 |
| chr22_+_40045451 | 0.13 |
ENST00000402203.5
|
TNRC6B
|
trinucleotide repeat containing adaptor 6B |
| chr14_+_66824439 | 0.13 |
ENST00000555456.1
|
GPHN
|
gephyrin |
| chr1_-_27374816 | 0.13 |
ENST00000270879.9
ENST00000354982.2 |
FCN3
|
ficolin 3 |
| chr2_+_202073249 | 0.13 |
ENST00000498697.3
|
KIAA2012
|
KIAA2012 |
| chr1_+_165827574 | 0.13 |
ENST00000367879.9
|
UCK2
|
uridine-cytidine kinase 2 |
| chr7_+_27242700 | 0.12 |
ENST00000222761.7
|
EVX1
|
even-skipped homeobox 1 |
| chr5_+_134525649 | 0.12 |
ENST00000282605.8
ENST00000681547.2 ENST00000361895.6 ENST00000402835.5 |
JADE2
|
jade family PHD finger 2 |
| chr4_-_185535498 | 0.12 |
ENST00000284767.12
ENST00000284771.7 ENST00000284770.10 ENST00000620787.5 ENST00000629667.2 |
PDLIM3
|
PDZ and LIM domain 3 |
| chr19_+_38789198 | 0.12 |
ENST00000314980.5
|
LGALS7B
|
galectin 7B |
| chr20_+_46029206 | 0.12 |
ENST00000243964.7
|
SLC12A5
|
solute carrier family 12 member 5 |
| chr6_+_36678699 | 0.12 |
ENST00000405375.5
ENST00000244741.10 ENST00000373711.3 |
CDKN1A
|
cyclin dependent kinase inhibitor 1A |
| chr20_+_46029165 | 0.11 |
ENST00000616201.4
ENST00000616202.4 ENST00000616933.4 ENST00000626937.2 |
SLC12A5
|
solute carrier family 12 member 5 |
| chr17_-_7329266 | 0.11 |
ENST00000571887.5
ENST00000315614.11 ENST00000399464.7 ENST00000570460.5 |
NEURL4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr19_-_1863497 | 0.11 |
ENST00000617223.1
ENST00000250916.6 |
KLF16
|
Kruppel like factor 16 |
| chr8_-_81483226 | 0.11 |
ENST00000256104.5
|
FABP4
|
fatty acid binding protein 4 |
| chr17_-_56833889 | 0.11 |
ENST00000397861.7
|
C17orf67
|
chromosome 17 open reading frame 67 |
| chr7_-_44325653 | 0.10 |
ENST00000440254.6
|
CAMK2B
|
calcium/calmodulin dependent protein kinase II beta |
| chr4_-_148444674 | 0.10 |
ENST00000344721.8
|
NR3C2
|
nuclear receptor subfamily 3 group C member 2 |
| chr15_+_89776326 | 0.10 |
ENST00000341735.5
|
MESP2
|
mesoderm posterior bHLH transcription factor 2 |
| chr8_-_98942557 | 0.10 |
ENST00000523601.5
|
STK3
|
serine/threonine kinase 3 |
| chr1_-_13165631 | 0.10 |
ENST00000323770.8
|
HNRNPCL4
|
heterogeneous nuclear ribonucleoprotein C like 4 |
| chr1_+_13061158 | 0.10 |
ENST00000681473.1
|
HNRNPCL3
|
heterogeneous nuclear ribonucleoprotein C like 3 |
| chr9_-_133149334 | 0.09 |
ENST00000393160.7
|
RALGDS
|
ral guanine nucleotide dissociation stimulator |
| chr1_-_11848345 | 0.09 |
ENST00000376476.1
|
NPPA
|
natriuretic peptide A |
| chr19_+_38899680 | 0.09 |
ENST00000576510.5
ENST00000392079.7 |
NFKBIB
|
NFKB inhibitor beta |
| chr9_-_111328520 | 0.09 |
ENST00000374428.1
|
OR2K2
|
olfactory receptor family 2 subfamily K member 2 |
| chr17_-_42181081 | 0.09 |
ENST00000607371.5
|
KCNH4
|
potassium voltage-gated channel subfamily H member 4 |
| chr5_-_34043205 | 0.09 |
ENST00000382065.8
ENST00000231338.7 |
C1QTNF3
|
C1q and TNF related 3 |
| chr5_-_138338325 | 0.09 |
ENST00000510119.1
ENST00000513970.5 |
CDC25C
|
cell division cycle 25C |
| chr17_+_7561899 | 0.09 |
ENST00000321337.12
|
SENP3
|
SUMO specific peptidase 3 |
| chr3_-_52409783 | 0.09 |
ENST00000470173.1
ENST00000296288.9 ENST00000460680.6 |
BAP1
|
BRCA1 associated protein 1 |
| chr5_-_132777229 | 0.09 |
ENST00000378721.8
ENST00000378719.7 ENST00000378701.5 |
SEPTIN8
|
septin 8 |
| chr8_+_102551583 | 0.08 |
ENST00000285402.4
|
ODF1
|
outer dense fiber of sperm tails 1 |
| chr17_-_42181116 | 0.08 |
ENST00000264661.4
|
KCNH4
|
potassium voltage-gated channel subfamily H member 4 |
| chr19_+_13906190 | 0.08 |
ENST00000318003.11
|
CC2D1A
|
coiled-coil and C2 domain containing 1A |
| chr1_+_151612001 | 0.08 |
ENST00000642376.1
ENST00000368843.8 ENST00000458013.7 |
SNX27
|
sorting nexin 27 |
| chr2_-_40452046 | 0.08 |
ENST00000406785.6
|
SLC8A1
|
solute carrier family 8 member A1 |
| chr15_-_82806054 | 0.08 |
ENST00000541889.1
ENST00000334574.12 ENST00000561368.1 |
FSD2
|
fibronectin type III and SPRY domain containing 2 |
| chr15_-_89751292 | 0.08 |
ENST00000300057.4
|
MESP1
|
mesoderm posterior bHLH transcription factor 1 |
| chr5_-_132777215 | 0.08 |
ENST00000458488.2
|
SEPTIN8
|
septin 8 |
| chr1_+_206507546 | 0.08 |
ENST00000580449.5
ENST00000581503.6 |
RASSF5
|
Ras association domain family member 5 |
| chr6_-_159745186 | 0.08 |
ENST00000537657.5
|
SOD2
|
superoxide dismutase 2 |
| chr2_-_199457931 | 0.08 |
ENST00000417098.6
|
SATB2
|
SATB homeobox 2 |
| chr7_+_27242796 | 0.08 |
ENST00000496902.7
|
EVX1
|
even-skipped homeobox 1 |
| chr19_+_13906255 | 0.08 |
ENST00000589606.5
|
CC2D1A
|
coiled-coil and C2 domain containing 1A |
| chrX_+_16786421 | 0.08 |
ENST00000398155.4
ENST00000380122.10 |
TXLNG
|
taxilin gamma |
| chrX_+_71254781 | 0.08 |
ENST00000677446.1
|
NONO
|
non-POU domain containing octamer binding |
| chr2_+_27496830 | 0.07 |
ENST00000264717.7
|
GCKR
|
glucokinase regulator |
| chr9_-_122213874 | 0.07 |
ENST00000482062.1
|
LHX6
|
LIM homeobox 6 |
| chr17_-_48604959 | 0.07 |
ENST00000225648.4
ENST00000484302.3 |
HOXB6
|
homeobox B6 |
| chr1_-_182672232 | 0.07 |
ENST00000508450.5
|
RGS8
|
regulator of G protein signaling 8 |
| chr7_-_44325617 | 0.07 |
ENST00000358707.7
ENST00000457475.5 |
CAMK2B
|
calcium/calmodulin dependent protein kinase II beta |
| chrX_+_123184153 | 0.07 |
ENST00000616590.4
|
GRIA3
|
glutamate ionotropic receptor AMPA type subunit 3 |
| chr12_+_93571832 | 0.07 |
ENST00000549887.1
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr10_+_92691813 | 0.07 |
ENST00000472590.6
|
HHEX
|
hematopoietically expressed homeobox |
| chr6_+_132552693 | 0.07 |
ENST00000275200.1
|
TAAR8
|
trace amine associated receptor 8 |
| chr12_-_102478539 | 0.07 |
ENST00000424202.6
|
IGF1
|
insulin like growth factor 1 |
| chr9_+_122519141 | 0.07 |
ENST00000340750.1
|
OR1J4
|
olfactory receptor family 1 subfamily J member 4 |
| chr9_-_122213903 | 0.07 |
ENST00000464484.3
|
LHX6
|
LIM homeobox 6 |
| chr1_-_112704921 | 0.07 |
ENST00000414971.1
ENST00000534717.5 |
RHOC
|
ras homolog family member C |
| chr5_-_180291967 | 0.07 |
ENST00000425491.6
ENST00000523583.1 ENST00000393360.7 ENST00000455781.5 ENST00000452135.7 ENST00000347470.8 ENST00000343111.10 |
MAPK9
|
mitogen-activated protein kinase 9 |
| chr7_+_143316105 | 0.07 |
ENST00000343257.7
ENST00000650516.1 |
CLCN1
|
chloride voltage-gated channel 1 |
| chr11_-_85719160 | 0.07 |
ENST00000389958.7
ENST00000527794.5 |
SYTL2
|
synaptotagmin like 2 |
| chr16_-_4345904 | 0.07 |
ENST00000571941.5
|
PAM16
|
presequence translocase associated motor 16 |
| chr11_-_85719111 | 0.07 |
ENST00000529581.5
ENST00000533577.1 |
SYTL2
|
synaptotagmin like 2 |
| chr2_+_206274643 | 0.07 |
ENST00000374423.9
ENST00000649650.1 |
ZDBF2
|
zinc finger DBF-type containing 2 |
| chr19_-_42412347 | 0.07 |
ENST00000601189.1
ENST00000599211.1 |
LIPE
|
lipase E, hormone sensitive type |
| chr21_-_30497160 | 0.07 |
ENST00000334058.3
|
KRTAP19-4
|
keratin associated protein 19-4 |
| chr9_-_14314567 | 0.07 |
ENST00000397579.6
|
NFIB
|
nuclear factor I B |
| chr19_-_54360949 | 0.07 |
ENST00000622064.1
|
LAIR1
|
leukocyte associated immunoglobulin like receptor 1 |
| chr2_+_206275002 | 0.06 |
ENST00000650097.1
ENST00000649525.1 ENST00000649998.1 |
ZDBF2
|
zinc finger DBF-type containing 2 |
| chr1_+_17580474 | 0.06 |
ENST00000375415.5
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor 10 like |
| chr15_+_40569290 | 0.06 |
ENST00000315616.12
ENST00000559271.1 ENST00000616318.1 |
RPUSD2
|
RNA pseudouridine synthase domain containing 2 |
| chr11_-_89065969 | 0.06 |
ENST00000305447.5
|
GRM5
|
glutamate metabotropic receptor 5 |
| chr1_-_11847772 | 0.06 |
ENST00000376480.7
ENST00000610706.1 |
NPPA
|
natriuretic peptide A |
| chr2_-_40453438 | 0.06 |
ENST00000455476.5
|
SLC8A1
|
solute carrier family 8 member A1 |
| chr20_+_2536573 | 0.06 |
ENST00000358864.2
|
TMC2
|
transmembrane channel like 2 |
| chr5_-_124744513 | 0.06 |
ENST00000504926.5
|
ZNF608
|
zinc finger protein 608 |
| chr1_-_28643005 | 0.06 |
ENST00000263974.4
ENST00000373824.9 ENST00000495422.2 |
TAF12
|
TATA-box binding protein associated factor 12 |
| chr9_-_14314519 | 0.06 |
ENST00000397581.6
|
NFIB
|
nuclear factor I B |
| chr2_-_40430257 | 0.06 |
ENST00000408028.6
ENST00000332839.8 ENST00000406391.2 ENST00000405901.7 |
SLC8A1
|
solute carrier family 8 member A1 |
| chr10_+_70052582 | 0.06 |
ENST00000676699.1
|
MACROH2A2
|
macroH2A.2 histone |
| chr10_-_97771954 | 0.06 |
ENST00000266066.4
|
SFRP5
|
secreted frizzled related protein 5 |
| chr12_-_90955172 | 0.06 |
ENST00000358859.3
|
CCER1
|
coiled-coil glutamate rich protein 1 |
| chr21_-_42350987 | 0.06 |
ENST00000291526.5
|
TFF2
|
trefoil factor 2 |
| chr7_+_2647703 | 0.06 |
ENST00000403167.5
|
TTYH3
|
tweety family member 3 |
| chr9_-_136746006 | 0.06 |
ENST00000476567.1
|
LCN6
|
lipocalin 6 |
| chrX_-_55489806 | 0.06 |
ENST00000500968.4
|
USP51
|
ubiquitin specific peptidase 51 |
| chr5_+_150660841 | 0.06 |
ENST00000297130.4
|
MYOZ3
|
myozenin 3 |
| chr5_-_173328407 | 0.06 |
ENST00000265087.9
|
STC2
|
stanniocalcin 2 |
| chr17_-_10804092 | 0.06 |
ENST00000581851.1
|
TMEM238L
|
transmembrane protein 238 like |
| chr17_+_78214286 | 0.06 |
ENST00000592734.5
ENST00000587746.5 |
BIRC5
|
baculoviral IAP repeat containing 5 |
| chr4_+_139301478 | 0.06 |
ENST00000296543.10
ENST00000398947.1 |
NAA15
|
N-alpha-acetyltransferase 15, NatA auxiliary subunit |
| chr16_+_2883228 | 0.06 |
ENST00000573965.1
ENST00000572006.1 |
FLYWCH2
|
FLYWCH family member 2 |
| chr3_-_109337572 | 0.06 |
ENST00000335658.6
|
DPPA4
|
developmental pluripotency associated 4 |
| chr20_+_1135178 | 0.06 |
ENST00000435720.5
|
PSMF1
|
proteasome inhibitor subunit 1 |
| chr20_+_1135217 | 0.06 |
ENST00000381898.5
|
PSMF1
|
proteasome inhibitor subunit 1 |
| chr17_+_27471999 | 0.06 |
ENST00000583370.5
ENST00000509603.6 ENST00000268763.10 ENST00000398988.7 |
KSR1
|
kinase suppressor of ras 1 |
| chr10_-_101695119 | 0.06 |
ENST00000331272.9
ENST00000664783.1 |
FBXW4
|
F-box and WD repeat domain containing 4 |
| chr4_-_151760977 | 0.06 |
ENST00000512306.5
ENST00000508611.1 ENST00000515812.5 ENST00000263985.11 |
GATB
|
glutamyl-tRNA amidotransferase subunit B |
| chr4_+_185209577 | 0.06 |
ENST00000652585.1
|
SNX25
|
sorting nexin 25 |
| chr19_-_15232399 | 0.05 |
ENST00000221730.8
|
EPHX3
|
epoxide hydrolase 3 |
| chr20_+_836052 | 0.05 |
ENST00000246100.3
|
FAM110A
|
family with sequence similarity 110 member A |
| chr1_+_147541491 | 0.05 |
ENST00000683836.1
ENST00000234739.8 |
BCL9
|
BCL9 transcription coactivator |
| chr7_-_139716980 | 0.05 |
ENST00000342645.7
|
HIPK2
|
homeodomain interacting protein kinase 2 |
| chr11_-_67508091 | 0.05 |
ENST00000531506.1
|
CDK2AP2
|
cyclin dependent kinase 2 associated protein 2 |
| chr12_+_93572664 | 0.05 |
ENST00000551556.2
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr7_-_99929620 | 0.05 |
ENST00000312891.3
|
GJC3
|
gap junction protein gamma 3 |
| chr11_-_796185 | 0.05 |
ENST00000533385.5
ENST00000528936.5 ENST00000629634.2 ENST00000625752.2 ENST00000528606.5 ENST00000320230.9 |
SLC25A22
|
solute carrier family 25 member 22 |
| chr2_+_74421721 | 0.05 |
ENST00000409791.5
ENST00000348227.4 |
WDR54
|
WD repeat domain 54 |
| chr8_-_143986425 | 0.05 |
ENST00000313059.9
ENST00000524918.5 ENST00000313028.12 ENST00000525773.5 |
PARP10
|
poly(ADP-ribose) polymerase family member 10 |
| chr19_+_13024917 | 0.05 |
ENST00000587260.1
|
NFIX
|
nuclear factor I X |
| chr1_-_161238163 | 0.05 |
ENST00000367982.8
|
NR1I3
|
nuclear receptor subfamily 1 group I member 3 |
| chr3_-_39154558 | 0.05 |
ENST00000514182.1
|
CSRNP1
|
cysteine and serine rich nuclear protein 1 |
| chr12_-_119803383 | 0.05 |
ENST00000392520.2
ENST00000678677.1 ENST00000679249.1 ENST00000676849.1 |
CIT
|
citron rho-interacting serine/threonine kinase |
| chr1_+_206507520 | 0.05 |
ENST00000579436.7
|
RASSF5
|
Ras association domain family member 5 |
| chr17_+_57256727 | 0.05 |
ENST00000675656.1
|
MSI2
|
musashi RNA binding protein 2 |
| chr2_-_113235443 | 0.05 |
ENST00000465084.1
|
PAX8
|
paired box 8 |
| chr15_-_82647503 | 0.05 |
ENST00000567678.1
ENST00000620182.4 |
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr1_+_200027605 | 0.05 |
ENST00000236914.7
|
NR5A2
|
nuclear receptor subfamily 5 group A member 2 |
| chr7_-_150978284 | 0.05 |
ENST00000262186.10
|
KCNH2
|
potassium voltage-gated channel subfamily H member 2 |
| chr4_-_139301204 | 0.05 |
ENST00000505036.5
ENST00000539002.5 ENST00000544855.5 |
NDUFC1
|
NADH:ubiquinone oxidoreductase subunit C1 |
| chr2_-_96857984 | 0.05 |
ENST00000393537.5
|
ANKRD39
|
ankyrin repeat domain 39 |
| chr17_+_44847874 | 0.05 |
ENST00000253410.3
|
HIGD1B
|
HIG1 hypoxia inducible domain family member 1B |
| chr1_-_112715469 | 0.05 |
ENST00000309276.10
|
PPM1J
|
protein phosphatase, Mg2+/Mn2+ dependent 1J |
| chr1_+_91952162 | 0.05 |
ENST00000402388.1
ENST00000680541.1 |
BRDT
|
bromodomain testis associated |
| chr20_+_11917859 | 0.05 |
ENST00000618296.4
ENST00000378226.7 |
BTBD3
|
BTB domain containing 3 |
| chr1_+_39081316 | 0.05 |
ENST00000484793.5
|
MACF1
|
microtubule actin crosslinking factor 1 |
| chr7_-_105679089 | 0.05 |
ENST00000477775.5
|
ATXN7L1
|
ataxin 7 like 1 |
| chr11_-_34513785 | 0.05 |
ENST00000257832.7
ENST00000429939.6 |
ELF5
|
E74 like ETS transcription factor 5 |
| chrX_+_76427666 | 0.05 |
ENST00000361470.4
|
MAGEE1
|
MAGE family member E1 |
| chr19_-_4558417 | 0.05 |
ENST00000586965.1
|
SEMA6B
|
semaphorin 6B |
| chr1_-_112715283 | 0.05 |
ENST00000464951.1
|
PPM1J
|
protein phosphatase, Mg2+/Mn2+ dependent 1J |
| chr4_-_75902444 | 0.05 |
ENST00000286719.12
|
PPEF2
|
protein phosphatase with EF-hand domain 2 |
| chr2_-_197675578 | 0.05 |
ENST00000295049.9
|
RFTN2
|
raftlin family member 2 |
| chr11_-_27722021 | 0.05 |
ENST00000314915.6
|
BDNF
|
brain derived neurotrophic factor |
| chr17_+_68291487 | 0.05 |
ENST00000621439.5
ENST00000452479.6 |
ARSG
|
arylsulfatase G |
| chr1_-_156816738 | 0.05 |
ENST00000368198.7
|
SH2D2A
|
SH2 domain containing 2A |
| chr2_-_151971750 | 0.05 |
ENST00000636598.1
|
CACNB4
|
calcium voltage-gated channel auxiliary subunit beta 4 |
| chr12_-_122227491 | 0.04 |
ENST00000475784.1
ENST00000645606.1 |
ENSG00000284934.2
|
novel protein |
| chr19_-_46471484 | 0.04 |
ENST00000313683.15
ENST00000602246.1 |
PNMA8A
|
PNMA family member 8A |
| chrX_-_132489842 | 0.04 |
ENST00000436215.5
|
MBNL3
|
muscleblind like splicing regulator 3 |
| chr1_+_150549734 | 0.04 |
ENST00000674043.1
ENST00000674058.1 |
ADAMTSL4
|
ADAMTS like 4 |
| chr19_-_46471407 | 0.04 |
ENST00000438932.2
|
PNMA8A
|
PNMA family member 8A |
| chr7_+_150405146 | 0.04 |
ENST00000498682.3
ENST00000641717.1 |
ENSG00000284691.1
|
novel zinc finger protein |
| chr2_-_74421560 | 0.04 |
ENST00000612891.4
ENST00000640331.1 ENST00000684111.1 |
C2orf81
|
chromosome 2 open reading frame 81 |
| chr22_+_35383106 | 0.04 |
ENST00000678411.1
|
HMOX1
|
heme oxygenase 1 |
| chr1_-_156816841 | 0.04 |
ENST00000368199.8
ENST00000392306.2 |
SH2D2A
|
SH2 domain containing 2A |
| chr6_+_54307856 | 0.04 |
ENST00000370869.7
|
TINAG
|
tubulointerstitial nephritis antigen |
| chr3_+_41194741 | 0.04 |
ENST00000643541.1
ENST00000426215.5 ENST00000645210.1 ENST00000646381.1 ENST00000405570.6 ENST00000642248.1 ENST00000433400.6 |
CTNNB1
|
catenin beta 1 |
| chr12_+_57755060 | 0.04 |
ENST00000266643.6
|
MARCHF9
|
membrane associated ring-CH-type finger 9 |
| chr12_-_84912705 | 0.04 |
ENST00000679933.1
ENST00000680260.1 ENST00000551010.2 ENST00000679453.1 ENST00000681281.1 |
SLC6A15
|
solute carrier family 6 member 15 |
| chr2_-_25878445 | 0.04 |
ENST00000336112.9
ENST00000435504.9 |
ASXL2
|
ASXL transcriptional regulator 2 |
| chr10_+_93073873 | 0.04 |
ENST00000224356.5
|
CYP26A1
|
cytochrome P450 family 26 subfamily A member 1 |
| chr11_-_85719045 | 0.04 |
ENST00000533057.6
ENST00000533892.5 |
SYTL2
|
synaptotagmin like 2 |
| chr2_-_216081759 | 0.04 |
ENST00000265322.8
|
PECR
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr17_-_42681840 | 0.04 |
ENST00000332438.4
|
CCR10
|
C-C motif chemokine receptor 10 |
| chr1_+_158355894 | 0.04 |
ENST00000368162.2
|
CD1E
|
CD1e molecule |
| chr11_-_34513750 | 0.04 |
ENST00000532417.1
|
ELF5
|
E74 like ETS transcription factor 5 |
| chr6_+_45422564 | 0.04 |
ENST00000625924.1
|
RUNX2
|
RUNX family transcription factor 2 |
| chr7_-_127392687 | 0.04 |
ENST00000393313.5
ENST00000619291.4 ENST00000265827.8 ENST00000434602.5 |
ZNF800
|
zinc finger protein 800 |
| chr10_+_93496599 | 0.04 |
ENST00000371485.8
|
CEP55
|
centrosomal protein 55 |
| chr17_+_49219503 | 0.04 |
ENST00000573347.5
|
ABI3
|
ABI family member 3 |
| chr8_-_108248700 | 0.04 |
ENST00000220849.10
ENST00000678937.1 ENST00000678901.1 ENST00000678243.1 ENST00000677674.1 ENST00000521297.2 ENST00000677272.1 ENST00000522352.6 ENST00000678042.1 ENST00000519627.2 ENST00000521440.6 ENST00000678773.1 ENST00000676698.1 ENST00000518345.2 ENST00000677447.1 ENST00000676548.1 ENST00000519030.6 ENST00000518442.5 |
EIF3E
|
eukaryotic translation initiation factor 3 subunit E |
| chr1_+_145719483 | 0.04 |
ENST00000369288.7
|
CD160
|
CD160 molecule |
| chr12_-_26125023 | 0.04 |
ENST00000242728.5
|
BHLHE41
|
basic helix-loop-helix family member e41 |
| chr9_-_76906041 | 0.04 |
ENST00000443509.6
ENST00000428286.5 ENST00000376713.3 |
PRUNE2
|
prune homolog 2 with BCH domain |
| chr14_-_52791597 | 0.04 |
ENST00000216410.8
ENST00000557604.1 |
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chrX_+_72069659 | 0.04 |
ENST00000631375.1
|
NHSL2
|
NHS like 2 |
| chr17_-_42276341 | 0.04 |
ENST00000293328.8
|
STAT5B
|
signal transducer and activator of transcription 5B |
| chr20_-_1329131 | 0.04 |
ENST00000360779.4
|
SDCBP2
|
syndecan binding protein 2 |
| chr14_+_78403686 | 0.04 |
ENST00000553631.1
ENST00000554719.5 |
NRXN3
|
neurexin 3 |
| chr22_+_31754862 | 0.04 |
ENST00000382111.6
ENST00000645407.1 ENST00000646701.1 |
DEPDC5
ENSG00000285404.1
|
DEP domain containing 5, GATOR1 subcomplex subunit novel protein, DEPDC5-YWHAH readthrough |
| chrX_-_134796256 | 0.04 |
ENST00000486347.5
|
PABIR2
|
PABIR family member 2 |
| chr4_+_1793285 | 0.04 |
ENST00000440486.8
ENST00000412135.7 ENST00000481110.7 ENST00000340107.8 |
FGFR3
|
fibroblast growth factor receptor 3 |
| chr17_+_7687416 | 0.04 |
ENST00000457584.6
|
WRAP53
|
WD repeat containing antisense to TP53 |
| chr1_-_110606009 | 0.04 |
ENST00000640774.2
ENST00000638616.2 |
KCNA2
|
potassium voltage-gated channel subfamily A member 2 |
| chr20_+_45881218 | 0.04 |
ENST00000372523.1
|
ZSWIM1
|
zinc finger SWIM-type containing 1 |
| chr6_+_42746958 | 0.04 |
ENST00000614467.4
|
BICRAL
|
BRD4 interacting chromatin remodeling complex associated protein like |
| chr2_+_201129318 | 0.04 |
ENST00000417748.1
|
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chr8_+_32647080 | 0.04 |
ENST00000520502.7
ENST00000523041.2 ENST00000650819.1 |
NRG1
|
neuregulin 1 |
| chr5_+_177133741 | 0.04 |
ENST00000439151.7
|
NSD1
|
nuclear receptor binding SET domain protein 1 |
| chr17_-_41065879 | 0.04 |
ENST00000394015.3
|
KRTAP2-4
|
keratin associated protein 2-4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX15_MGA
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0061011 | hepatic duct development(GO:0061011) |
| 0.1 | 0.2 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.1 | 0.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 0.2 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.1 | GO:0018315 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.3 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.2 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.0 | 0.2 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.2 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.1 | GO:0060913 | cardiac cell fate determination(GO:0060913) negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.2 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:0003064 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.2 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.0 | 0.2 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.2 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.0 | GO:0001207 | histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.0 | 0.1 | GO:0003070 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.0 | GO:0031550 | positive regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031550) |
| 0.0 | 0.1 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:1904073 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.0 | 0.0 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.0 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.1 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.2 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.0 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.1 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.0 | GO:0032831 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 0.0 | 0.2 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.1 | GO:0001712 | ectoderm formation(GO:0001705) ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.0 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.0 | 0.1 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.1 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.1 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.0 | GO:0097252 | negative regulation of helicase activity(GO:0051097) oligodendrocyte apoptotic process(GO:0097252) |
| 0.0 | 0.3 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.1 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.0 | 0.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.0 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.1 | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.0 | 0.0 | GO:1903173 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.2 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.0 | 0.1 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.1 | GO:0008940 | nitrate reductase activity(GO:0008940) molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.1 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.2 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.2 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.0 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.0 | 0.1 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.1 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.0 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.0 | GO:0019166 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.0 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.0 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.1 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.0 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 0.1 | GO:0086077 | gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0004882 | androgen receptor activity(GO:0004882) |