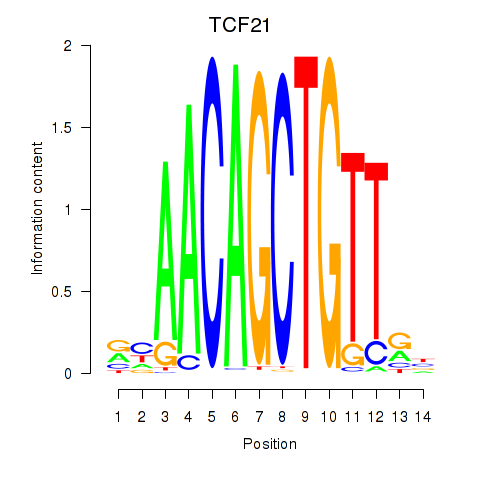
|
chr12_+_77830886
Show fit
|
0.30 |
ENST00000397909.7
ENST00000549464.5
|
NAV3
|
neuron navigator 3
|
|
chr4_+_113292925
Show fit
|
0.28 |
ENST00000673353.1
ENST00000505342.6
ENST00000672915.1
ENST00000509550.5
|
ANK2
|
ankyrin 2
|
|
chr4_+_113292838
Show fit
|
0.28 |
ENST00000672411.1
ENST00000673231.1
|
ANK2
|
ankyrin 2
|
|
chr11_+_71527267
Show fit
|
0.22 |
ENST00000398536.6
|
KRTAP5-7
|
keratin associated protein 5-7
|
|
chr17_+_7438267
Show fit
|
0.19 |
ENST00000575235.5
|
FGF11
|
fibroblast growth factor 11
|
|
chr1_-_243163310
Show fit
|
0.17 |
ENST00000492145.1
ENST00000490813.5
ENST00000464936.5
|
CEP170
|
centrosomal protein 170
|
|
chr11_-_59866478
Show fit
|
0.17 |
ENST00000257264.4
|
TCN1
|
transcobalamin 1
|
|
chr16_+_56336727
Show fit
|
0.16 |
ENST00000568375.2
|
GNAO1
|
G protein subunit alpha o1
|
|
chr4_+_122923067
Show fit
|
0.15 |
ENST00000675612.1
ENST00000274008.5
|
SPATA5
|
spermatogenesis associated 5
|
|
chr9_+_27109393
Show fit
|
0.15 |
ENST00000406359.8
|
TEK
|
TEK receptor tyrosine kinase
|
|
chr15_+_40695423
Show fit
|
0.15 |
ENST00000526763.6
ENST00000532743.6
|
RAD51
|
RAD51 recombinase
|
|
chr18_-_49849827
Show fit
|
0.14 |
ENST00000592688.1
|
MYO5B
|
myosin VB
|
|
chr4_-_46909206
Show fit
|
0.14 |
ENST00000396533.5
|
COX7B2
|
cytochrome c oxidase subunit 7B2
|
|
chr3_-_127590707
Show fit
|
0.14 |
ENST00000296210.11
ENST00000355552.8
|
TPRA1
|
transmembrane protein adipocyte associated 1
|
|
chr3_+_3799424
Show fit
|
0.14 |
ENST00000319331.4
|
LRRN1
|
leucine rich repeat neuronal 1
|
|
chr11_-_796185
Show fit
|
0.14 |
ENST00000533385.5
ENST00000528936.5
ENST00000629634.2
ENST00000625752.2
ENST00000528606.5
ENST00000320230.9
|
SLC25A22
|
solute carrier family 25 member 22
|
|
chrX_+_70290077
Show fit
|
0.13 |
ENST00000374403.4
|
KIF4A
|
kinesin family member 4A
|
|
chrX_-_109625161
Show fit
|
0.13 |
ENST00000372101.3
|
KCNE5
|
potassium voltage-gated channel subfamily E regulatory subunit 5
|
|
chr3_+_148865288
Show fit
|
0.13 |
ENST00000296046.4
|
CPA3
|
carboxypeptidase A3
|
|
chr2_-_224947030
Show fit
|
0.13 |
ENST00000409592.7
|
DOCK10
|
dedicator of cytokinesis 10
|
|
chrX_-_104013708
Show fit
|
0.13 |
ENST00000217926.7
|
H2BW1
|
H2B.W histone 1
|
|
chr5_-_16936231
Show fit
|
0.11 |
ENST00000507288.1
ENST00000274203.13
ENST00000513610.6
|
MYO10
|
myosin X
|
|
chr7_-_93226449
Show fit
|
0.11 |
ENST00000394468.7
ENST00000453812.2
|
HEPACAM2
|
HEPACAM family member 2
|
|
chr19_+_6531018
Show fit
|
0.11 |
ENST00000245817.5
|
TNFSF9
|
TNF superfamily member 9
|
|
chr10_+_35126923
Show fit
|
0.11 |
ENST00000374726.7
|
CREM
|
cAMP responsive element modulator
|
|
chr12_-_81598332
Show fit
|
0.10 |
ENST00000443686.7
|
PPFIA2
|
PTPRF interacting protein alpha 2
|
|
chr4_-_46909235
Show fit
|
0.10 |
ENST00000505102.1
ENST00000355591.8
|
COX7B2
|
cytochrome c oxidase subunit 7B2
|
|
chr5_-_179623098
Show fit
|
0.10 |
ENST00000630639.1
ENST00000329433.10
ENST00000510411.5
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1
|
|
chr16_-_81220370
Show fit
|
0.10 |
ENST00000337114.8
|
PKD1L2
|
polycystin 1 like 2 (gene/pseudogene)
|
|
chr19_-_45076465
Show fit
|
0.10 |
ENST00000303809.7
|
ZNF296
|
zinc finger protein 296
|
|
chr15_+_89243945
Show fit
|
0.10 |
ENST00000674831.1
ENST00000300027.12
ENST00000567891.5
ENST00000310775.12
ENST00000676003.1
ENST00000564920.5
ENST00000565255.5
ENST00000567996.5
ENST00000563250.5
|
FANCI
|
FA complementation group I
|
|
chr4_-_152679984
Show fit
|
0.10 |
ENST00000304385.8
ENST00000504064.1
|
TMEM154
|
transmembrane protein 154
|
|
chr1_-_44141631
Show fit
|
0.10 |
ENST00000634670.1
|
KLF18
|
Kruppel like factor 18
|
|
chr1_+_22636577
Show fit
|
0.10 |
ENST00000374642.8
ENST00000438241.1
|
C1QA
|
complement C1q A chain
|
|
chr8_+_25459190
Show fit
|
0.10 |
ENST00000380665.3
ENST00000330560.8
|
CDCA2
|
cell division cycle associated 2
|
|
chr5_+_892844
Show fit
|
0.10 |
ENST00000166345.8
|
TRIP13
|
thyroid hormone receptor interactor 13
|
|
chr16_-_5097742
Show fit
|
0.10 |
ENST00000587133.1
ENST00000458008.8
ENST00000427587.9
|
EEF2KMT
|
eukaryotic elongation factor 2 lysine methyltransferase
|
|
chr1_-_149917826
Show fit
|
0.09 |
ENST00000369145.1
ENST00000369146.8
|
SV2A
|
synaptic vesicle glycoprotein 2A
|
|
chr13_-_49936486
Show fit
|
0.09 |
ENST00000378195.6
|
SPRYD7
|
SPRY domain containing 7
|
|
chr22_+_49853801
Show fit
|
0.09 |
ENST00000216268.6
|
ZBED4
|
zinc finger BED-type containing 4
|
|
chr19_-_45076504
Show fit
|
0.09 |
ENST00000622376.1
|
ZNF296
|
zinc finger protein 296
|
|
chr21_-_44261854
Show fit
|
0.09 |
ENST00000431166.1
|
DNMT3L
|
DNA methyltransferase 3 like
|
|
chr2_-_121736911
Show fit
|
0.09 |
ENST00000451734.1
|
NIFK
|
nucleolar protein interacting with the FHA domain of MKI67
|
|
chr17_-_7178116
Show fit
|
0.09 |
ENST00000380920.8
|
ASGR1
|
asialoglycoprotein receptor 1
|
|
chr2_-_233854566
Show fit
|
0.09 |
ENST00000432087.5
ENST00000441687.5
ENST00000414924.5
|
HJURP
|
Holliday junction recognition protein
|
|
chr20_+_17699942
Show fit
|
0.09 |
ENST00000427254.1
ENST00000377805.7
|
BANF2
|
BANF family member 2
|
|
chr13_-_49936298
Show fit
|
0.09 |
ENST00000613924.1
ENST00000361840.8
|
SPRYD7
|
SPRY domain containing 7
|
|
chr11_-_89922245
Show fit
|
0.09 |
ENST00000420869.3
|
TRIM49D1
|
tripartite motif containing 49D1
|
|
chr10_+_92834594
Show fit
|
0.08 |
ENST00000371552.8
|
EXOC6
|
exocyst complex component 6
|
|
chr16_-_8936633
Show fit
|
0.08 |
ENST00000381886.8
|
USP7
|
ubiquitin specific peptidase 7
|
|
chr2_+_232697299
Show fit
|
0.08 |
ENST00000476995.5
ENST00000427233.5
ENST00000629305.2
ENST00000428883.5
ENST00000456491.5
ENST00000409480.5
ENST00000492910.5
ENST00000464402.5
ENST00000490612.5
ENST00000475359.6
ENST00000421433.5
ENST00000425040.5
ENST00000430720.5
ENST00000409547.5
ENST00000373563.9
ENST00000423659.5
ENST00000409196.7
ENST00000488734.5
ENST00000409451.7
ENST00000429187.5
ENST00000440945.5
|
GIGYF2
|
GRB10 interacting GYF protein 2
|
|
chr4_+_15374541
Show fit
|
0.08 |
ENST00000382383.7
ENST00000429690.5
|
C1QTNF7
|
C1q and TNF related 7
|
|
chr19_+_22832284
Show fit
|
0.08 |
ENST00000600766.3
|
ZNF723
|
zinc finger protein 723
|
|
chr6_-_31680377
Show fit
|
0.08 |
ENST00000383237.4
|
LY6G5C
|
lymphocyte antigen 6 family member G5C
|
|
chr5_-_11589019
Show fit
|
0.08 |
ENST00000511377.5
|
CTNND2
|
catenin delta 2
|
|
chr5_+_69167216
Show fit
|
0.08 |
ENST00000508407.5
ENST00000505500.5
|
CCNB1
|
cyclin B1
|
|
chr10_-_12042771
Show fit
|
0.08 |
ENST00000357604.10
|
UPF2
|
UPF2 regulator of nonsense mediated mRNA decay
|
|
chr8_-_140718661
Show fit
|
0.08 |
ENST00000430260.6
|
PTK2
|
protein tyrosine kinase 2
|
|
chr2_-_233854506
Show fit
|
0.08 |
ENST00000411486.7
|
HJURP
|
Holliday junction recognition protein
|
|
chr20_+_31739260
Show fit
|
0.08 |
ENST00000340513.4
ENST00000300403.11
|
TPX2
|
TPX2 microtubule nucleation factor
|
|
chr6_+_10528326
Show fit
|
0.08 |
ENST00000379597.7
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2 (I blood group)
|
|
chr17_-_44947637
Show fit
|
0.08 |
ENST00000587309.5
ENST00000593135.6
|
KIF18B
|
kinesin family member 18B
|
|
chr18_+_3247778
Show fit
|
0.08 |
ENST00000217652.8
ENST00000578611.5
ENST00000583449.1
|
MYL12A
|
myosin light chain 12A
|
|
chr2_+_68157877
Show fit
|
0.07 |
ENST00000263657.7
|
PNO1
|
partner of NOB1 homolog
|
|
chr22_+_20917398
Show fit
|
0.07 |
ENST00000354336.8
|
CRKL
|
CRK like proto-oncogene, adaptor protein
|
|
chr5_-_60700094
Show fit
|
0.07 |
ENST00000453022.6
ENST00000265036.10
|
DEPDC1B
|
DEP domain containing 1B
|
|
chr8_-_66667138
Show fit
|
0.07 |
ENST00000310421.5
|
VCPIP1
|
valosin containing protein interacting protein 1
|
|
chr3_-_33659097
Show fit
|
0.07 |
ENST00000461133.8
ENST00000496954.2
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr12_-_6851245
Show fit
|
0.07 |
ENST00000540683.1
ENST00000229265.10
ENST00000535406.5
ENST00000422785.7
ENST00000538862.7
|
CDCA3
|
cell division cycle associated 3
|
|
chr8_+_66043413
Show fit
|
0.07 |
ENST00000522619.1
|
DNAJC5B
|
DnaJ heat shock protein family (Hsp40) member C5 beta
|
|
chr1_+_23743462
Show fit
|
0.07 |
ENST00000609199.1
|
ELOA
|
elongin A
|
|
chr11_-_88175432
Show fit
|
0.07 |
ENST00000531138.1
ENST00000526372.1
ENST00000243662.11
|
RAB38
|
RAB38, member RAS oncogene family
|
|
chr17_+_6444441
Show fit
|
0.07 |
ENST00000250056.12
ENST00000571373.5
ENST00000570337.6
ENST00000572595.6
ENST00000572447.6
ENST00000576056.5
|
PIMREG
|
PICALM interacting mitotic regulator
|
|
chr1_-_24415035
Show fit
|
0.07 |
ENST00000374409.5
|
STPG1
|
sperm tail PG-rich repeat containing 1
|
|
chr9_+_122375286
Show fit
|
0.07 |
ENST00000373698.7
|
PTGS1
|
prostaglandin-endoperoxide synthase 1
|
|
chr13_+_41061481
Show fit
|
0.07 |
ENST00000379487.5
|
WBP4
|
WW domain binding protein 4
|
|
chr3_+_63278277
Show fit
|
0.07 |
ENST00000478300.6
ENST00000450542.6
|
SYNPR
|
synaptoporin
|
|
chr2_-_17800195
Show fit
|
0.07 |
ENST00000402989.5
ENST00000428868.1
|
SMC6
|
structural maintenance of chromosomes 6
|
|
chr1_-_225889143
Show fit
|
0.07 |
ENST00000272134.5
|
LEFTY1
|
left-right determination factor 1
|
|
chr18_+_23949847
Show fit
|
0.07 |
ENST00000588004.1
|
LAMA3
|
laminin subunit alpha 3
|
|
chr5_-_115296610
Show fit
|
0.07 |
ENST00000379611.10
ENST00000506442.5
|
CCDC112
|
coiled-coil domain containing 112
|
|
chr1_-_23531206
Show fit
|
0.07 |
ENST00000361729.3
|
E2F2
|
E2F transcription factor 2
|
|
chr2_+_219434825
Show fit
|
0.07 |
ENST00000312358.12
|
SPEG
|
striated muscle enriched protein kinase
|
|
chr4_-_88231322
Show fit
|
0.07 |
ENST00000515655.5
|
ABCG2
|
ATP binding cassette subfamily G member 2 (Junior blood group)
|
|
chr9_-_19102887
Show fit
|
0.07 |
ENST00000380502.8
|
HAUS6
|
HAUS augmin like complex subunit 6
|
|
chr7_-_100100716
Show fit
|
0.07 |
ENST00000354230.7
ENST00000425308.5
|
MCM7
|
minichromosome maintenance complex component 7
|
|
chr1_-_228416626
Show fit
|
0.07 |
ENST00000355586.4
ENST00000520264.1
ENST00000479800.1
ENST00000295033.7
|
TRIM17
|
tripartite motif containing 17
|
|
chr13_-_52159844
Show fit
|
0.07 |
ENST00000611833.4
|
NEK3
|
NIMA related kinase 3
|
|
chr13_-_52159529
Show fit
|
0.07 |
ENST00000618534.4
ENST00000550841.1
|
NEK3
|
NIMA related kinase 3
|
|
chr7_+_150737382
Show fit
|
0.07 |
ENST00000358647.5
|
GIMAP5
|
GTPase, IMAP family member 5
|
|
chr6_-_25830557
Show fit
|
0.07 |
ENST00000468082.1
|
SLC17A1
|
solute carrier family 17 member 1
|
|
chr5_+_108924585
Show fit
|
0.07 |
ENST00000618353.1
|
FER
|
FER tyrosine kinase
|
|
chr6_-_11382247
Show fit
|
0.07 |
ENST00000397378.7
ENST00000513989.5
ENST00000508546.5
ENST00000504387.5
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr9_+_27109135
Show fit
|
0.07 |
ENST00000519097.5
ENST00000615002.4
|
TEK
|
TEK receptor tyrosine kinase
|
|
chr12_-_112108743
Show fit
|
0.07 |
ENST00000547133.1
ENST00000261745.9
|
NAA25
|
N-alpha-acetyltransferase 25, NatB auxiliary subunit
|
|
chr11_+_89924064
Show fit
|
0.07 |
ENST00000623787.3
|
TRIM49D2
|
tripartite motif containing 49D2
|
|
chr1_-_30757767
Show fit
|
0.06 |
ENST00000294507.4
|
LAPTM5
|
lysosomal protein transmembrane 5
|
|
chr21_-_44262216
Show fit
|
0.06 |
ENST00000270172.7
|
DNMT3L
|
DNA methyltransferase 3 like
|
|
chr22_-_22508702
Show fit
|
0.06 |
ENST00000626650.3
|
ZNF280B
|
zinc finger protein 280B
|
|
chr1_-_23800402
Show fit
|
0.06 |
ENST00000374497.7
ENST00000425913.5
|
GALE
|
UDP-galactose-4-epimerase
|
|
chr13_-_52159559
Show fit
|
0.06 |
ENST00000620675.4
ENST00000610828.5
|
NEK3
|
NIMA related kinase 3
|
|
chr4_+_95051671
Show fit
|
0.06 |
ENST00000440890.7
|
BMPR1B
|
bone morphogenetic protein receptor type 1B
|
|
chr1_+_22001654
Show fit
|
0.06 |
ENST00000290122.8
|
CELA3A
|
chymotrypsin like elastase 3A
|
|
chr17_+_82217929
Show fit
|
0.06 |
ENST00000580098.6
|
SLC16A3
|
solute carrier family 16 member 3
|
|
chr19_-_17075038
Show fit
|
0.06 |
ENST00000593360.1
|
HAUS8
|
HAUS augmin like complex subunit 8
|
|
chr11_+_75151095
Show fit
|
0.06 |
ENST00000289575.10
ENST00000525650.5
ENST00000454962.6
|
SLCO2B1
|
solute carrier organic anion transporter family member 2B1
|
|
chr10_+_88664439
Show fit
|
0.06 |
ENST00000394375.7
ENST00000608620.5
ENST00000238983.9
ENST00000355843.2
|
LIPF
|
lipase F, gastric type
|
|
chr5_+_69167117
Show fit
|
0.06 |
ENST00000506572.5
ENST00000256442.10
|
CCNB1
|
cyclin B1
|
|
chr19_-_17075418
Show fit
|
0.06 |
ENST00000253669.10
|
HAUS8
|
HAUS augmin like complex subunit 8
|
|
chr16_-_31428325
Show fit
|
0.06 |
ENST00000287490.5
|
COX6A2
|
cytochrome c oxidase subunit 6A2
|
|
chr5_-_179623659
Show fit
|
0.06 |
ENST00000519056.5
ENST00000506721.5
ENST00000503105.5
ENST00000504348.5
ENST00000508103.5
ENST00000510431.5
ENST00000515158.5
ENST00000393432.8
ENST00000442819.6
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1
|
|
chr1_+_172452885
Show fit
|
0.06 |
ENST00000367725.4
|
C1orf105
|
chromosome 1 open reading frame 105
|
|
chr14_-_52791462
Show fit
|
0.06 |
ENST00000650397.1
ENST00000554230.5
|
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1
|
|
chr17_-_41149823
Show fit
|
0.06 |
ENST00000343246.6
|
KRTAP4-5
|
keratin associated protein 4-5
|
|
chr18_+_59139864
Show fit
|
0.06 |
ENST00000509791.7
ENST00000587834.6
ENST00000588875.2
|
SEC11C
|
SEC11 homolog C, signal peptidase complex subunit
|
|
chr12_+_121888751
Show fit
|
0.06 |
ENST00000261817.6
ENST00000538613.5
ENST00000542602.1
ENST00000541212.6
|
PSMD9
|
proteasome 26S subunit, non-ATPase 9
|
|
chr2_+_232697362
Show fit
|
0.06 |
ENST00000482666.5
ENST00000483164.5
ENST00000490229.5
ENST00000464805.5
ENST00000489328.1
|
GIGYF2
|
GRB10 interacting GYF protein 2
|
|
chr7_-_155510158
Show fit
|
0.06 |
ENST00000682997.1
|
CNPY1
|
canopy FGF signaling regulator 1
|
|
chr6_+_30617825
Show fit
|
0.06 |
ENST00000259873.5
|
MRPS18B
|
mitochondrial ribosomal protein S18B
|
|
chr6_-_73452124
Show fit
|
0.06 |
ENST00000680833.1
|
CGAS
|
cyclic GMP-AMP synthase
|
|
chr3_-_33659441
Show fit
|
0.06 |
ENST00000650653.1
ENST00000480013.6
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr3_+_52211442
Show fit
|
0.06 |
ENST00000459884.1
|
ALAS1
|
5'-aminolevulinate synthase 1
|
|
chr12_+_57434778
Show fit
|
0.06 |
ENST00000309668.3
|
INHBC
|
inhibin subunit beta C
|
|
chr6_-_31684040
Show fit
|
0.06 |
ENST00000375863.7
|
LY6G5C
|
lymphocyte antigen 6 family member G5C
|
|
chr9_-_120926752
Show fit
|
0.06 |
ENST00000373887.8
|
TRAF1
|
TNF receptor associated factor 1
|
|
chr1_+_212565334
Show fit
|
0.06 |
ENST00000366981.8
ENST00000366987.6
|
ATF3
|
activating transcription factor 3
|
|
chr14_-_22819721
Show fit
|
0.06 |
ENST00000554517.5
ENST00000285850.11
ENST00000397529.6
ENST00000555702.5
|
SLC7A7
|
solute carrier family 7 member 7
|
|
chr11_+_827545
Show fit
|
0.05 |
ENST00000528542.6
|
CRACR2B
|
calcium release activated channel regulator 2B
|
|
chr11_+_34105582
Show fit
|
0.05 |
ENST00000531159.6
ENST00000257829.8
|
NAT10
|
N-acetyltransferase 10
|
|
chrX_+_149774068
Show fit
|
0.05 |
ENST00000370416.4
|
HSFX1
|
heat shock transcription factor family, X-linked 1
|
|
chr7_-_41703062
Show fit
|
0.05 |
ENST00000242208.5
|
INHBA
|
inhibin subunit beta A
|
|
chr8_+_85438850
Show fit
|
0.05 |
ENST00000285381.3
|
CA3
|
carbonic anhydrase 3
|
|
chr22_-_21952827
Show fit
|
0.05 |
ENST00000397495.8
ENST00000263212.10
|
PPM1F
|
protein phosphatase, Mg2+/Mn2+ dependent 1F
|
|
chr12_+_8697875
Show fit
|
0.05 |
ENST00000357529.7
|
RIMKLB
|
ribosomal modification protein rimK like family member B
|
|
chr20_+_836052
Show fit
|
0.05 |
ENST00000246100.3
|
FAM110A
|
family with sequence similarity 110 member A
|
|
chr12_-_109021015
Show fit
|
0.05 |
ENST00000546618.2
ENST00000610966.5
|
SVOP
|
SV2 related protein
|
|
chr12_+_8697945
Show fit
|
0.05 |
ENST00000535829.6
|
RIMKLB
|
ribosomal modification protein rimK like family member B
|
|
chr14_+_67533282
Show fit
|
0.05 |
ENST00000329153.10
|
PLEKHH1
|
pleckstrin homology, MyTH4 and FERM domain containing H1
|
|
chr17_-_41424583
Show fit
|
0.05 |
ENST00000225550.4
|
KRT37
|
keratin 37
|
|
chr1_-_228416841
Show fit
|
0.05 |
ENST00000366698.7
ENST00000457345.2
|
TRIM17
|
tripartite motif containing 17
|
|
chr15_+_92904447
Show fit
|
0.05 |
ENST00000626782.2
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr7_+_142800957
Show fit
|
0.05 |
ENST00000466254.1
|
TRBC2
|
T cell receptor beta constant 2
|
|
chr11_+_827913
Show fit
|
0.05 |
ENST00000525077.2
|
CRACR2B
|
calcium release activated channel regulator 2B
|
|
chr21_-_44261884
Show fit
|
0.05 |
ENST00000628202.3
|
DNMT3L
|
DNA methyltransferase 3 like
|
|
chr8_-_20183127
Show fit
|
0.05 |
ENST00000276373.10
ENST00000519026.5
ENST00000440926.3
ENST00000437980.3
|
SLC18A1
|
solute carrier family 18 member A1
|
|
chr5_+_126631680
Show fit
|
0.05 |
ENST00000357147.4
|
TEX43
|
testis expressed 43
|
|
chr16_-_70289480
Show fit
|
0.05 |
ENST00000261772.13
ENST00000675953.1
ENST00000675691.1
ENST00000675133.1
ENST00000674512.1
ENST00000675045.1
ENST00000675853.1
ENST00000565361.3
ENST00000674963.1
ENST00000674691.1
|
AARS1
|
alanyl-tRNA synthetase 1
|
|
chr5_+_122312229
Show fit
|
0.05 |
ENST00000261368.13
|
SNCAIP
|
synuclein alpha interacting protein
|
|
chr1_-_165445088
Show fit
|
0.05 |
ENST00000359842.10
|
RXRG
|
retinoid X receptor gamma
|
|
chr2_+_27663880
Show fit
|
0.05 |
ENST00000618046.4
ENST00000613517.4
|
SLC4A1AP
|
solute carrier family 4 member 1 adaptor protein
|
|
chr2_-_88979016
Show fit
|
0.05 |
ENST00000390247.2
|
IGKV3-7
|
immunoglobulin kappa variable 3-7 (non-functional)
|
|
chr8_-_20183090
Show fit
|
0.05 |
ENST00000265808.11
ENST00000522513.5
|
SLC18A1
|
solute carrier family 18 member A1
|
|
chr6_+_97010193
Show fit
|
0.05 |
ENST00000620278.1
|
KLHL32
|
kelch like family member 32
|
|
chr21_-_41512090
Show fit
|
0.05 |
ENST00000679263.1
|
TMPRSS2
|
transmembrane serine protease 2
|
|
chr1_+_1033987
Show fit
|
0.05 |
ENST00000651234.1
ENST00000652369.1
|
AGRN
|
agrin
|
|
chr5_-_36001006
Show fit
|
0.05 |
ENST00000625798.2
|
UGT3A1
|
UDP glycosyltransferase family 3 member A1
|
|
chr19_-_51905004
Show fit
|
0.05 |
ENST00000600738.5
ENST00000354957.8
ENST00000595418.5
ENST00000599530.1
|
ZNF649
|
zinc finger protein 649
|
|
chr3_+_8733779
Show fit
|
0.05 |
ENST00000343849.3
ENST00000397368.2
|
CAV3
|
caveolin 3
|
|
chr22_+_21015027
Show fit
|
0.04 |
ENST00000413302.6
ENST00000401443.5
|
P2RX6
|
purinergic receptor P2X 6
|
|
chr2_-_218671975
Show fit
|
0.04 |
ENST00000295704.7
|
RNF25
|
ring finger protein 25
|
|
chr9_+_27109200
Show fit
|
0.04 |
ENST00000380036.10
|
TEK
|
TEK receptor tyrosine kinase
|
|
chr12_+_8989612
Show fit
|
0.04 |
ENST00000266551.8
|
KLRG1
|
killer cell lectin like receptor G1
|
|
chr15_+_65550819
Show fit
|
0.04 |
ENST00000569894.5
|
HACD3
|
3-hydroxyacyl-CoA dehydratase 3
|
|
chr2_-_55049184
Show fit
|
0.04 |
ENST00000357376.7
|
RTN4
|
reticulon 4
|
|
chr17_+_19411220
Show fit
|
0.04 |
ENST00000461366.2
|
RNF112
|
ring finger protein 112
|
|
chr1_+_145927105
Show fit
|
0.04 |
ENST00000437797.5
ENST00000601726.3
ENST00000599626.5
ENST00000599147.5
ENST00000595494.5
ENST00000595518.5
ENST00000597144.5
ENST00000599469.5
ENST00000598354.5
ENST00000598103.5
ENST00000600340.5
ENST00000630257.2
ENST00000625258.1
|
LIX1L-AS1
ENSG00000280778.1
|
LIX1L antisense RNA 1
novel protein, lncRNA-POLR3GL readthrough
|
|
chr2_-_121736863
Show fit
|
0.04 |
ENST00000285814.9
|
NIFK
|
nucleolar protein interacting with the FHA domain of MKI67
|
|
chr2_-_189762755
Show fit
|
0.04 |
ENST00000520350.1
ENST00000521630.1
ENST00000264151.10
ENST00000517895.5
|
OSGEPL1
|
O-sialoglycoprotein endopeptidase like 1
|
|
chr3_-_52835011
Show fit
|
0.04 |
ENST00000446157.3
|
MUSTN1
|
musculoskeletal, embryonic nuclear protein 1
|
|
chr9_+_99821876
Show fit
|
0.04 |
ENST00000395097.7
|
NR4A3
|
nuclear receptor subfamily 4 group A member 3
|
|
chr1_+_186296267
Show fit
|
0.04 |
ENST00000533951.5
ENST00000367482.8
ENST00000635041.1
ENST00000367483.8
ENST00000367485.4
ENST00000445192.7
|
PRG4
|
proteoglycan 4
|
|
chr19_-_50719761
Show fit
|
0.04 |
ENST00000293441.6
|
SHANK1
|
SH3 and multiple ankyrin repeat domains 1
|
|
chr3_-_185254028
Show fit
|
0.04 |
ENST00000440662.1
ENST00000456310.5
ENST00000231887.8
|
EHHADH
|
enoyl-CoA hydratase and 3-hydroxyacyl CoA dehydrogenase
|
|
chr5_-_892765
Show fit
|
0.04 |
ENST00000467963.6
|
BRD9
|
bromodomain containing 9
|
|
chr5_-_180802790
Show fit
|
0.04 |
ENST00000504671.1
ENST00000507384.1
ENST00000307826.5
|
MGAT1
|
alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase
|
|
chr5_-_151093566
Show fit
|
0.04 |
ENST00000521001.1
|
TNIP1
|
TNFAIP3 interacting protein 1
|
|
chr1_-_165445220
Show fit
|
0.04 |
ENST00000619224.1
|
RXRG
|
retinoid X receptor gamma
|
|
chr2_+_90234809
Show fit
|
0.04 |
ENST00000443397.5
|
IGKV3D-7
|
immunoglobulin kappa variable 3D-7
|
|
chr3_+_142723999
Show fit
|
0.04 |
ENST00000476941.6
ENST00000273482.10
|
TRPC1
|
transient receptor potential cation channel subfamily C member 1
|
|
chr11_+_75151055
Show fit
|
0.04 |
ENST00000532236.5
|
SLCO2B1
|
solute carrier organic anion transporter family member 2B1
|
|
chr1_+_222928415
Show fit
|
0.04 |
ENST00000284476.7
|
DISP1
|
dispatched RND transporter family member 1
|
|
chr16_+_81645352
Show fit
|
0.04 |
ENST00000398040.8
|
CMIP
|
c-Maf inducing protein
|
|
chr2_+_87338511
Show fit
|
0.04 |
ENST00000421835.2
|
IGKV3OR2-268
|
immunoglobulin kappa variable 3/OR2-268 (non-functional)
|
|
chr3_-_52834901
Show fit
|
0.04 |
ENST00000486659.5
|
MUSTN1
|
musculoskeletal, embryonic nuclear protein 1
|
|
chr21_-_32279012
Show fit
|
0.04 |
ENST00000290130.4
|
MIS18A
|
MIS18 kinetochore protein A
|
|
chr18_+_58221535
Show fit
|
0.04 |
ENST00000431212.6
ENST00000586268.5
ENST00000587190.5
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase
|
|
chr10_-_95441015
Show fit
|
0.04 |
ENST00000371241.5
ENST00000354106.7
ENST00000371239.5
ENST00000361941.7
ENST00000277982.9
ENST00000371245.7
|
SORBS1
|
sorbin and SH3 domain containing 1
|
|
chr1_-_160711803
Show fit
|
0.04 |
ENST00000368045.3
ENST00000368046.8
ENST00000613788.1
|
CD48
|
CD48 molecule
|
|
chr2_+_6877768
Show fit
|
0.04 |
ENST00000382040.4
|
RSAD2
|
radical S-adenosyl methionine domain containing 2
|
|
chr12_-_81598360
Show fit
|
0.04 |
ENST00000333447.11
ENST00000407050.8
|
PPFIA2
|
PTPRF interacting protein alpha 2
|
|
chr3_+_101779182
Show fit
|
0.04 |
ENST00000495842.5
ENST00000273347.10
|
NXPE3
|
neurexophilin and PC-esterase domain family member 3
|
|
chr15_-_23039560
Show fit
|
0.04 |
ENST00000615383.5
ENST00000620435.4
|
TUBGCP5
|
tubulin gamma complex associated protein 5
|
|
chr3_+_101779425
Show fit
|
0.04 |
ENST00000491511.6
|
NXPE3
|
neurexophilin and PC-esterase domain family member 3
|
|
chr12_+_16347665
Show fit
|
0.04 |
ENST00000535309.5
ENST00000540056.5
ENST00000396209.5
ENST00000540126.5
|
MGST1
|
microsomal glutathione S-transferase 1
|
|
chr7_+_150716603
Show fit
|
0.04 |
ENST00000307194.6
ENST00000611999.4
|
GIMAP1
GIMAP1-GIMAP5
|
GTPase, IMAP family member 1
GIMAP1-GIMAP5 readthrough
|
|
chr9_-_110999458
Show fit
|
0.04 |
ENST00000374430.6
|
LPAR1
|
lysophosphatidic acid receptor 1
|
|
chr19_+_18340629
Show fit
|
0.04 |
ENST00000597431.2
|
PGPEP1
|
pyroglutamyl-peptidase I
|
|
chr16_-_4273014
Show fit
|
0.04 |
ENST00000204517.11
|
TFAP4
|
transcription factor AP-4
|
|
chr2_+_32357009
Show fit
|
0.04 |
ENST00000421745.6
|
BIRC6
|
baculoviral IAP repeat containing 6
|
|
chr1_-_226408045
Show fit
|
0.03 |
ENST00000366794.10
ENST00000677203.1
|
PARP1
|
poly(ADP-ribose) polymerase 1
|