Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
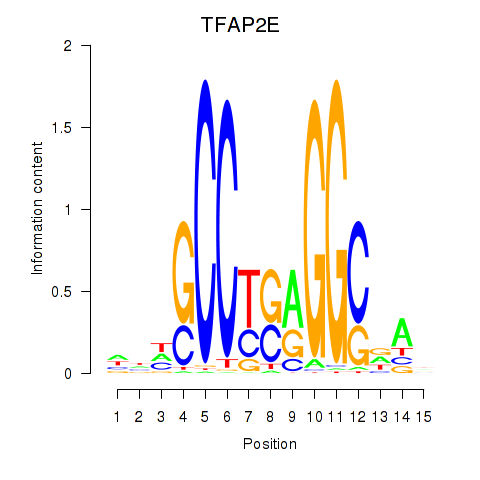
Results for TFAP2E
Z-value: 0.29
Transcription factors associated with TFAP2E
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TFAP2E
|
ENSG00000116819.9 | transcription factor AP-2 epsilon |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TFAP2E | hg38_v1_chr1_+_35573308_35573319 | 0.20 | 6.4e-01 | Click! |
Activity profile of TFAP2E motif
Sorted Z-values of TFAP2E motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_54173151 | 0.18 |
ENST00000619895.5
|
TMC4
|
transmembrane channel like 4 |
| chr19_-_54173190 | 0.16 |
ENST00000617472.4
|
TMC4
|
transmembrane channel like 4 |
| chr19_-_43465596 | 0.16 |
ENST00000244333.4
|
LYPD3
|
LY6/PLAUR domain containing 3 |
| chr19_-_38773432 | 0.12 |
ENST00000599035.1
ENST00000378626.5 |
LGALS7
|
galectin 7 |
| chr19_+_45001430 | 0.11 |
ENST00000625761.2
ENST00000505236.1 ENST00000221452.13 |
RELB
|
RELB proto-oncogene, NF-kB subunit |
| chr3_+_50155305 | 0.11 |
ENST00000002829.8
ENST00000426511.5 |
SEMA3F
|
semaphorin 3F |
| chr17_-_41612757 | 0.11 |
ENST00000301653.9
ENST00000593067.1 |
KRT16
|
keratin 16 |
| chr1_-_32336224 | 0.10 |
ENST00000329421.8
|
MARCKSL1
|
MARCKS like 1 |
| chr20_+_6767678 | 0.10 |
ENST00000378827.5
|
BMP2
|
bone morphogenetic protein 2 |
| chr22_+_39994926 | 0.10 |
ENST00000333407.11
|
FAM83F
|
family with sequence similarity 83 member F |
| chr11_+_118607579 | 0.09 |
ENST00000530708.4
|
PHLDB1
|
pleckstrin homology like domain family B member 1 |
| chr18_-_76495191 | 0.07 |
ENST00000443185.7
|
ZNF516
|
zinc finger protein 516 |
| chr1_-_85048437 | 0.07 |
ENST00000341115.8
ENST00000370587.5 ENST00000370589.7 |
MCOLN3
|
mucolipin TRP cation channel 3 |
| chr6_+_13272709 | 0.07 |
ENST00000379335.8
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr22_+_31248402 | 0.07 |
ENST00000333611.8
ENST00000340552.4 |
LIMK2
|
LIM domain kinase 2 |
| chr22_+_31212207 | 0.06 |
ENST00000406516.5
ENST00000331728.9 |
LIMK2
|
LIM domain kinase 2 |
| chr2_+_186694007 | 0.06 |
ENST00000304698.10
|
FAM171B
|
family with sequence similarity 171 member B |
| chr8_-_126557691 | 0.06 |
ENST00000652209.1
|
LRATD2
|
LRAT domain containing 2 |
| chr15_-_65422894 | 0.06 |
ENST00000352385.3
|
IGDCC4
|
immunoglobulin superfamily DCC subclass member 4 |
| chr9_+_135714425 | 0.05 |
ENST00000473941.5
ENST00000486577.6 |
KCNT1
|
potassium sodium-activated channel subfamily T member 1 |
| chr2_+_6917404 | 0.05 |
ENST00000320892.11
|
RNF144A
|
ring finger protein 144A |
| chr3_+_52779916 | 0.05 |
ENST00000537050.5
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr3_-_55487295 | 0.05 |
ENST00000264634.9
|
WNT5A
|
Wnt family member 5A |
| chr19_+_35704540 | 0.04 |
ENST00000392197.7
ENST00000426659.6 |
ZBTB32
|
zinc finger and BTB domain containing 32 |
| chr2_+_190649062 | 0.04 |
ENST00000409581.5
ENST00000337386.10 |
NAB1
|
NGFI-A binding protein 1 |
| chr2_+_74834113 | 0.04 |
ENST00000290573.7
|
HK2
|
hexokinase 2 |
| chr13_-_75482151 | 0.04 |
ENST00000377636.8
|
TBC1D4
|
TBC1 domain family member 4 |
| chr2_+_38666059 | 0.04 |
ENST00000272252.10
ENST00000410063.5 |
GALM
|
galactose mutarotase |
| chr1_-_11805924 | 0.04 |
ENST00000418034.1
|
MTHFR
|
methylenetetrahydrofolate reductase |
| chr13_-_75482114 | 0.04 |
ENST00000377625.6
ENST00000431480.6 |
TBC1D4
|
TBC1 domain family member 4 |
| chr12_-_31324129 | 0.03 |
ENST00000454658.6
|
SINHCAF
|
SIN3-HDAC complex associated factor |
| chr19_+_43576800 | 0.03 |
ENST00000612042.4
|
PINLYP
|
phospholipase A2 inhibitor and LY6/PLAUR domain containing |
| chr1_-_40862354 | 0.03 |
ENST00000372638.4
|
CITED4
|
Cbp/p300 interacting transactivator with Glu/Asp rich carboxy-terminal domain 4 |
| chr7_+_151085858 | 0.03 |
ENST00000463381.5
|
AGAP3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr1_-_11805949 | 0.03 |
ENST00000376590.9
|
MTHFR
|
methylenetetrahydrofolate reductase |
| chrX_-_124963768 | 0.03 |
ENST00000371130.7
ENST00000422452.2 |
TENM1
|
teneurin transmembrane protein 1 |
| chr7_+_148590760 | 0.03 |
ENST00000307003.3
|
C7orf33
|
chromosome 7 open reading frame 33 |
| chr15_+_84817346 | 0.03 |
ENST00000258888.6
|
ALPK3
|
alpha kinase 3 |
| chr13_+_19958708 | 0.03 |
ENST00000382874.6
ENST00000610343.5 |
ZMYM2
|
zinc finger MYM-type containing 2 |
| chr4_+_6909775 | 0.03 |
ENST00000409757.9
|
TBC1D14
|
TBC1 domain family member 14 |
| chrX_+_153724847 | 0.03 |
ENST00000218104.6
|
ABCD1
|
ATP binding cassette subfamily D member 1 |
| chr4_+_6909444 | 0.03 |
ENST00000448507.5
|
TBC1D14
|
TBC1 domain family member 14 |
| chr19_-_42255119 | 0.03 |
ENST00000222329.9
ENST00000594664.1 |
ERF
ENSG00000268643.2
|
ETS2 repressor factor novel protein |
| chr11_-_66289007 | 0.03 |
ENST00000431556.6
ENST00000528575.1 |
YIF1A
|
Yip1 interacting factor homolog A, membrane trafficking protein |
| chr17_+_39667964 | 0.03 |
ENST00000394246.1
|
PNMT
|
phenylethanolamine N-methyltransferase |
| chr7_-_45088888 | 0.03 |
ENST00000490531.3
|
NACAD
|
NAC alpha domain containing |
| chr19_-_35490456 | 0.03 |
ENST00000338897.4
ENST00000484218.6 |
KRTDAP
|
keratinocyte differentiation associated protein |
| chr6_+_7107941 | 0.03 |
ENST00000379938.7
ENST00000467782.5 ENST00000334984.10 ENST00000349384.10 |
RREB1
|
ras responsive element binding protein 1 |
| chr20_+_19212624 | 0.03 |
ENST00000328041.11
|
SLC24A3
|
solute carrier family 24 member 3 |
| chr17_+_39668447 | 0.03 |
ENST00000269582.3
ENST00000581428.1 |
PNMT
|
phenylethanolamine N-methyltransferase |
| chr1_-_113812254 | 0.03 |
ENST00000616024.4
ENST00000615321.1 |
RSBN1
|
round spermatid basic protein 1 |
| chr2_+_186694076 | 0.03 |
ENST00000612606.1
|
FAM171B
|
family with sequence similarity 171 member B |
| chr1_-_43453902 | 0.03 |
ENST00000372430.9
ENST00000372432.5 ENST00000583037.5 |
HYI
|
hydroxypyruvate isomerase (putative) |
| chr3_+_4493340 | 0.03 |
ENST00000357086.10
ENST00000354582.12 ENST00000649015.2 ENST00000467056.6 |
ITPR1
|
inositol 1,4,5-trisphosphate receptor type 1 |
| chr1_-_43453716 | 0.02 |
ENST00000372433.5
|
HYI
|
hydroxypyruvate isomerase (putative) |
| chr17_-_44915486 | 0.02 |
ENST00000638281.1
ENST00000588316.1 ENST00000588735.3 ENST00000639277.1 ENST00000253408.11 ENST00000435360.8 ENST00000586793.6 ENST00000588037.1 ENST00000592320.6 |
GFAP
|
glial fibrillary acidic protein |
| chr3_+_49554436 | 0.02 |
ENST00000296452.5
|
BSN
|
bassoon presynaptic cytomatrix protein |
| chr2_-_177264686 | 0.02 |
ENST00000397062.8
ENST00000430047.1 |
NFE2L2
|
nuclear factor, erythroid 2 like 2 |
| chr1_-_11805977 | 0.02 |
ENST00000376486.3
|
MTHFR
|
methylenetetrahydrofolate reductase |
| chr1_-_43453792 | 0.02 |
ENST00000372434.5
ENST00000486909.1 |
HYI
|
hydroxypyruvate isomerase (putative) |
| chr19_+_49157780 | 0.02 |
ENST00000599628.5
ENST00000252826.10 ENST00000427978.6 |
TRPM4
|
transient receptor potential cation channel subfamily M member 4 |
| chr19_+_4969105 | 0.02 |
ENST00000611640.4
ENST00000159111.9 ENST00000588337.5 ENST00000381759.8 |
KDM4B
|
lysine demethylase 4B |
| chr14_+_103385450 | 0.02 |
ENST00000416682.6
|
MARK3
|
microtubule affinity regulating kinase 3 |
| chr10_-_110919146 | 0.02 |
ENST00000652396.1
ENST00000423273.5 ENST00000436562.1 ENST00000651952.1 ENST00000448814.7 ENST00000652400.1 ENST00000447005.5 ENST00000454061.5 |
BBIP1
|
BBSome interacting protein 1 |
| chr14_-_25049889 | 0.02 |
ENST00000419632.6
ENST00000396700.5 |
STXBP6
|
syntaxin binding protein 6 |
| chr10_-_124092445 | 0.02 |
ENST00000346248.7
|
CHST15
|
carbohydrate sulfotransferase 15 |
| chr19_+_8390316 | 0.02 |
ENST00000328024.11
ENST00000601897.1 ENST00000594216.1 |
RAB11B
|
RAB11B, member RAS oncogene family |
| chr11_-_61295289 | 0.02 |
ENST00000335613.10
|
VWCE
|
von Willebrand factor C and EGF domains |
| chr1_+_213989691 | 0.02 |
ENST00000607425.1
|
PROX1
|
prospero homeobox 1 |
| chr2_-_74529670 | 0.02 |
ENST00000377526.4
|
AUP1
|
AUP1 lipid droplet regulating VLDL assembly factor |
| chr3_+_4493442 | 0.02 |
ENST00000456211.8
ENST00000443694.5 ENST00000648266.1 |
ITPR1
|
inositol 1,4,5-trisphosphate receptor type 1 |
| chr1_-_113812448 | 0.02 |
ENST00000612242.4
ENST00000261441.9 |
RSBN1
|
round spermatid basic protein 1 |
| chr8_+_124450806 | 0.02 |
ENST00000328599.4
|
TRMT12
|
tRNA methyltransferase 12 homolog |
| chr14_+_103385374 | 0.02 |
ENST00000678179.1
ENST00000676938.1 ENST00000678619.1 ENST00000440884.7 ENST00000560417.6 ENST00000679330.1 ENST00000556744.2 ENST00000676897.1 ENST00000677560.1 ENST00000561314.6 ENST00000677829.1 ENST00000677133.1 ENST00000676645.1 ENST00000678175.1 ENST00000429436.7 ENST00000677360.1 ENST00000678237.1 ENST00000677347.1 ENST00000677432.1 |
MARK3
|
microtubule affinity regulating kinase 3 |
| chr13_+_19958760 | 0.02 |
ENST00000382871.3
|
ZMYM2
|
zinc finger MYM-type containing 2 |
| chr8_-_102655707 | 0.02 |
ENST00000285407.11
|
KLF10
|
Kruppel like factor 10 |
| chr14_+_103385506 | 0.02 |
ENST00000303622.13
|
MARK3
|
microtubule affinity regulating kinase 3 |
| chr17_+_42780592 | 0.02 |
ENST00000246914.10
|
WNK4
|
WNK lysine deficient protein kinase 4 |
| chr16_-_381935 | 0.02 |
ENST00000431232.7
ENST00000250930.7 |
PGAP6
|
post-glycosylphosphatidylinositol attachment to proteins 6 |
| chr2_-_240892007 | 0.02 |
ENST00000402775.6
ENST00000307486.12 |
MAB21L4
|
mab-21 like 4 |
| chr14_+_24368020 | 0.02 |
ENST00000554050.5
ENST00000554903.1 ENST00000250373.9 ENST00000554779.1 ENST00000553708.5 |
NFATC4
|
nuclear factor of activated T cells 4 |
| chr11_+_57805541 | 0.02 |
ENST00000683201.1
ENST00000683769.1 |
CTNND1
|
catenin delta 1 |
| chr6_+_42564060 | 0.02 |
ENST00000372903.6
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr20_+_24469623 | 0.02 |
ENST00000376862.4
|
SYNDIG1
|
synapse differentiation inducing 1 |
| chr11_-_35419213 | 0.02 |
ENST00000642171.1
ENST00000644050.1 ENST00000643134.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr14_-_25050111 | 0.02 |
ENST00000323944.9
|
STXBP6
|
syntaxin binding protein 6 |
| chrX_+_132023294 | 0.02 |
ENST00000481105.5
ENST00000354719.10 ENST00000394334.7 ENST00000394335.6 |
STK26
|
serine/threonine kinase 26 |
| chr11_-_27506751 | 0.02 |
ENST00000278193.7
ENST00000524596.1 |
LIN7C
|
lin-7 homolog C, crumbs cell polarity complex component |
| chr6_+_31586835 | 0.02 |
ENST00000211921.11
|
LST1
|
leukocyte specific transcript 1 |
| chr6_-_87702221 | 0.02 |
ENST00000257787.6
|
AKIRIN2
|
akirin 2 |
| chr2_+_218859794 | 0.01 |
ENST00000233948.4
|
WNT6
|
Wnt family member 6 |
| chr9_-_122213903 | 0.01 |
ENST00000464484.3
|
LHX6
|
LIM homeobox 6 |
| chr17_-_68601357 | 0.01 |
ENST00000592554.2
|
FAM20A
|
FAM20A golgi associated secretory pathway pseudokinase |
| chr11_-_66289125 | 0.01 |
ENST00000471387.6
ENST00000376901.9 ENST00000359461.10 |
YIF1A
|
Yip1 interacting factor homolog A, membrane trafficking protein |
| chr9_-_122213874 | 0.01 |
ENST00000482062.1
|
LHX6
|
LIM homeobox 6 |
| chr11_-_35419098 | 0.01 |
ENST00000606205.6
ENST00000645303.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr5_-_151924824 | 0.01 |
ENST00000455880.2
|
GLRA1
|
glycine receptor alpha 1 |
| chr5_-_151924846 | 0.01 |
ENST00000274576.9
|
GLRA1
|
glycine receptor alpha 1 |
| chr19_+_13118235 | 0.01 |
ENST00000292431.5
|
NACC1
|
nucleus accumbens associated 1 |
| chr11_-_35420017 | 0.01 |
ENST00000643000.1
ENST00000646099.1 ENST00000647372.1 ENST00000642578.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr1_+_33256479 | 0.01 |
ENST00000539719.6
ENST00000483388.5 |
ZNF362
|
zinc finger protein 362 |
| chr7_+_29194757 | 0.01 |
ENST00000222792.11
|
CHN2
|
chimerin 2 |
| chr10_-_110918934 | 0.01 |
ENST00000605742.5
|
BBIP1
|
BBSome interacting protein 1 |
| chr11_-_35419462 | 0.01 |
ENST00000643522.1
|
SLC1A2
|
solute carrier family 1 member 2 |
| chr7_-_100468063 | 0.01 |
ENST00000423266.5
ENST00000456330.1 |
TSC22D4
|
TSC22 domain family member 4 |
| chr3_+_156142962 | 0.01 |
ENST00000471742.5
|
KCNAB1
|
potassium voltage-gated channel subfamily A member regulatory beta subunit 1 |
| chr19_+_50432885 | 0.01 |
ENST00000357701.6
|
MYBPC2
|
myosin binding protein C2 |
| chr10_-_131981948 | 0.01 |
ENST00000633835.1
|
BNIP3
|
BCL2 interacting protein 3 |
| chr3_+_122795039 | 0.01 |
ENST00000261038.6
|
SLC49A4
|
solute carrier family 49 member 4 |
| chr1_+_3069160 | 0.01 |
ENST00000511072.5
|
PRDM16
|
PR/SET domain 16 |
| chr1_+_22563460 | 0.01 |
ENST00000166244.8
ENST00000374644.8 |
EPHA8
|
EPH receptor A8 |
| chr4_-_152679984 | 0.01 |
ENST00000304385.8
ENST00000504064.1 |
TMEM154
|
transmembrane protein 154 |
| chr1_+_3069195 | 0.01 |
ENST00000378391.6
ENST00000270722.10 ENST00000514189.5 |
PRDM16
|
PR/SET domain 16 |
| chr6_+_31586859 | 0.01 |
ENST00000433492.5
|
LST1
|
leukocyte specific transcript 1 |
| chr3_+_4493471 | 0.01 |
ENST00000544951.6
ENST00000650294.1 |
ITPR1
|
inositol 1,4,5-trisphosphate receptor type 1 |
| chr3_+_53161120 | 0.01 |
ENST00000394729.6
ENST00000330452.8 ENST00000652449.1 |
PRKCD
|
protein kinase C delta |
| chrX_-_49079702 | 0.01 |
ENST00000636049.1
ENST00000474053.6 ENST00000635003.1 |
WDR45
|
WD repeat domain 45 |
| chr16_-_75433391 | 0.01 |
ENST00000566254.1
ENST00000283882.4 |
CFDP1
|
craniofacial development protein 1 |
| chr17_-_76726753 | 0.01 |
ENST00000617192.4
|
JMJD6
|
jumonji domain containing 6, arginine demethylase and lysine hydroxylase |
| chr9_+_126326809 | 0.01 |
ENST00000361171.8
ENST00000489637.3 |
MVB12B
|
multivesicular body subunit 12B |
| chr2_-_37324826 | 0.01 |
ENST00000234179.8
|
PRKD3
|
protein kinase D3 |
| chr7_-_32495238 | 0.01 |
ENST00000409952.3
ENST00000409909.7 |
LSM5
|
LSM5 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr13_-_30465923 | 0.01 |
ENST00000341423.10
ENST00000326004.4 |
HMGB1
|
high mobility group box 1 |
| chr11_-_47848467 | 0.00 |
ENST00000378460.6
|
NUP160
|
nucleoporin 160 |
| chr6_+_42563981 | 0.00 |
ENST00000372899.6
ENST00000372901.2 |
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr11_-_47848313 | 0.00 |
ENST00000530326.5
ENST00000528071.5 |
NUP160
|
nucleoporin 160 |
| chr14_+_20745880 | 0.00 |
ENST00000326842.3
|
EDDM3A
|
epididymal protein 3A |
| chr22_-_38700655 | 0.00 |
ENST00000216039.9
|
JOSD1
|
Josephin domain containing 1 |
| chr1_-_224434750 | 0.00 |
ENST00000414423.9
ENST00000678917.1 ENST00000678555.1 ENST00000678307.1 |
WDR26
|
WD repeat domain 26 |
| chr7_+_73433761 | 0.00 |
ENST00000344575.5
|
FZD9
|
frizzled class receptor 9 |
| chr5_-_180071708 | 0.00 |
ENST00000522208.6
ENST00000521389.6 |
RNF130
|
ring finger protein 130 |
| chr9_+_133636355 | 0.00 |
ENST00000393056.8
|
DBH
|
dopamine beta-hydroxylase |
| chr12_-_14567714 | 0.00 |
ENST00000240617.10
|
PLBD1
|
phospholipase B domain containing 1 |
| chr6_-_18264475 | 0.00 |
ENST00000515742.2
ENST00000651624.1 ENST00000507591.2 ENST00000652689.1 ENST00000244776.11 ENST00000503715.5 |
DEK
|
DEK proto-oncogene |
| chr11_+_3855629 | 0.00 |
ENST00000526596.2
ENST00000300737.8 ENST00000616714.4 |
STIM1
|
stromal interaction molecule 1 |
| chr4_+_153466324 | 0.00 |
ENST00000409663.7
ENST00000409959.8 |
TMEM131L
|
transmembrane 131 like |
| chr7_-_140924900 | 0.00 |
ENST00000646891.1
ENST00000644969.2 |
BRAF
|
B-Raf proto-oncogene, serine/threonine kinase |
| chr10_-_124092390 | 0.00 |
ENST00000628426.1
|
CHST15
|
carbohydrate sulfotransferase 15 |
| chr11_+_118607598 | 0.00 |
ENST00000600882.6
ENST00000356063.9 |
PHLDB1
|
pleckstrin homology like domain family B member 1 |
| chr12_+_67269328 | 0.00 |
ENST00000545606.6
|
CAND1
|
cullin associated and neddylation dissociated 1 |
| chr7_+_130070518 | 0.00 |
ENST00000335420.10
ENST00000463413.1 |
KLHDC10
|
kelch domain containing 10 |
| chr9_+_35605234 | 0.00 |
ENST00000336395.6
|
TESK1
|
testis associated actin remodelling kinase 1 |
| chr13_+_99981775 | 0.00 |
ENST00000376335.8
|
ZIC2
|
Zic family member 2 |
| chrX_+_116436599 | 0.00 |
ENST00000598581.3
|
SLC6A14
|
solute carrier family 6 member 14 |
| chr11_-_65857007 | 0.00 |
ENST00000527344.5
|
CFL1
|
cofilin 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TFAP2E
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0051040 | regulation of calcium-independent cell-cell adhesion(GO:0051040) corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.0 | 0.1 | GO:0042418 | epinephrine biosynthetic process(GO:0042418) |
| 0.0 | 0.1 | GO:0036484 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.0 | 0.1 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.0 | GO:1904933 | cardiac right atrium morphogenesis(GO:0003213) mediolateral intercalation(GO:0060031) lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) planar cell polarity pathway involved in gastrula mediolateral intercalation(GO:0060775) non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) regulation of cell proliferation in midbrain(GO:1904933) |
| 0.0 | 0.0 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 0.1 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0008903 | hydroxypyruvate isomerase activity(GO:0008903) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |