Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
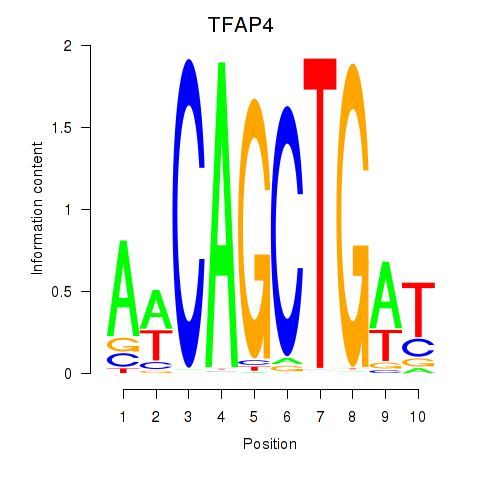
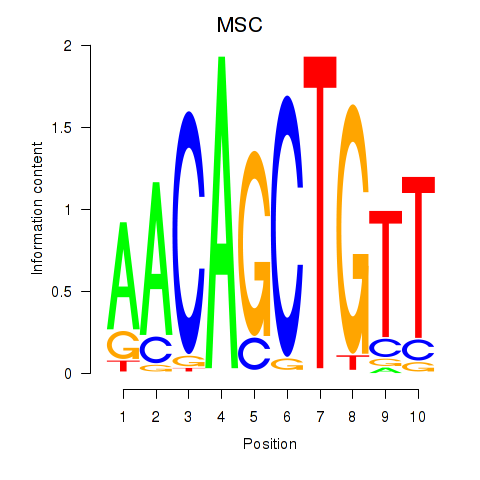
Results for TFAP4_MSC
Z-value: 0.95
Transcription factors associated with TFAP4_MSC
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TFAP4
|
ENSG00000090447.12 | transcription factor AP-4 |
|
MSC
|
ENSG00000178860.9 | musculin |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MSC | hg38_v1_chr8_-_71844402_71844429 | 0.82 | 1.3e-02 | Click! |
| TFAP4 | hg38_v1_chr16_-_4273014_4273065 | -0.66 | 7.8e-02 | Click! |
Activity profile of TFAP4_MSC motif
Sorted Z-values of TFAP4_MSC motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_55722857 | 1.15 |
ENST00000424486.3
|
TMEM100
|
transmembrane protein 100 |
| chr13_+_101452629 | 0.98 |
ENST00000622834.4
ENST00000545560.6 ENST00000376180.8 |
ITGBL1
|
integrin subunit beta like 1 |
| chr11_-_111912871 | 0.96 |
ENST00000528628.5
|
CRYAB
|
crystallin alpha B |
| chr13_+_101452569 | 0.94 |
ENST00000618057.4
|
ITGBL1
|
integrin subunit beta like 1 |
| chr5_+_176810552 | 0.92 |
ENST00000329542.9
|
UNC5A
|
unc-5 netrin receptor A |
| chr2_-_224947030 | 0.92 |
ENST00000409592.7
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr20_+_34704336 | 0.89 |
ENST00000374809.6
ENST00000374810.8 ENST00000451665.5 |
TP53INP2
|
tumor protein p53 inducible nuclear protein 2 |
| chr6_-_31729260 | 0.86 |
ENST00000375789.7
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr6_-_31729785 | 0.81 |
ENST00000416410.6
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr8_-_94949350 | 0.79 |
ENST00000448464.6
ENST00000342697.5 |
TP53INP1
|
tumor protein p53 inducible nuclear protein 1 |
| chr6_-_31729478 | 0.78 |
ENST00000436437.2
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chrX_-_107775951 | 0.74 |
ENST00000315660.8
ENST00000372384.6 ENST00000502650.1 ENST00000506724.1 |
TSC22D3
|
TSC22 domain family member 3 |
| chr5_+_95731300 | 0.72 |
ENST00000379982.8
|
RHOBTB3
|
Rho related BTB domain containing 3 |
| chr17_-_7241787 | 0.59 |
ENST00000577035.5
|
GABARAP
|
GABA type A receptor-associated protein |
| chr9_-_120477354 | 0.58 |
ENST00000416449.5
|
CDK5RAP2
|
CDK5 regulatory subunit associated protein 2 |
| chr2_-_213151590 | 0.54 |
ENST00000374319.8
ENST00000457361.5 ENST00000451136.6 ENST00000434687.6 |
IKZF2
|
IKAROS family zinc finger 2 |
| chr19_+_18097763 | 0.53 |
ENST00000262811.10
|
MAST3
|
microtubule associated serine/threonine kinase 3 |
| chr1_-_33182030 | 0.52 |
ENST00000291416.10
|
TRIM62
|
tripartite motif containing 62 |
| chr8_-_71362054 | 0.51 |
ENST00000340726.8
|
EYA1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr11_-_70662197 | 0.51 |
ENST00000409161.5
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr8_-_71361860 | 0.50 |
ENST00000303824.11
ENST00000645451.1 |
EYA1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr6_-_31728877 | 0.50 |
ENST00000437288.5
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr2_+_148021404 | 0.50 |
ENST00000638043.2
|
MBD5
|
methyl-CpG binding domain protein 5 |
| chr8_-_13276491 | 0.48 |
ENST00000512044.6
|
DLC1
|
DLC1 Rho GTPase activating protein |
| chr11_-_70661762 | 0.48 |
ENST00000357171.7
ENST00000412252.5 ENST00000449833.6 ENST00000338508.8 |
SHANK2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr2_-_174764407 | 0.46 |
ENST00000409219.5
ENST00000409542.5 |
CHRNA1
|
cholinergic receptor nicotinic alpha 1 subunit |
| chr12_-_53727428 | 0.46 |
ENST00000548263.5
ENST00000430117.6 ENST00000549173.5 ENST00000550804.6 ENST00000551900.5 ENST00000546619.5 ENST00000548177.5 ENST00000549349.5 |
CALCOCO1
|
calcium binding and coiled-coil domain 1 |
| chr15_-_52652031 | 0.46 |
ENST00000546305.6
|
FAM214A
|
family with sequence similarity 214 member A |
| chr17_-_55732074 | 0.45 |
ENST00000575734.5
|
TMEM100
|
transmembrane protein 100 |
| chr12_-_53727476 | 0.45 |
ENST00000549784.5
ENST00000262059.8 |
CALCOCO1
|
calcium binding and coiled-coil domain 1 |
| chr6_-_117425905 | 0.44 |
ENST00000368507.8
|
ROS1
|
ROS proto-oncogene 1, receptor tyrosine kinase |
| chr6_-_117425855 | 0.44 |
ENST00000368508.7
|
ROS1
|
ROS proto-oncogene 1, receptor tyrosine kinase |
| chr11_-_1763894 | 0.44 |
ENST00000637915.1
ENST00000637815.2 ENST00000236671.7 ENST00000636571.1 ENST00000438213.6 ENST00000637387.1 ENST00000636843.1 ENST00000636397.1 ENST00000636615.1 |
CTSD
ENSG00000250644.3
|
cathepsin D novel protein |
| chr13_-_42992165 | 0.43 |
ENST00000398762.7
ENST00000313640.11 ENST00000313624.12 |
EPSTI1
|
epithelial stromal interaction 1 |
| chr22_+_30607145 | 0.43 |
ENST00000405742.7
|
TCN2
|
transcobalamin 2 |
| chr7_-_137846860 | 0.43 |
ENST00000288490.9
ENST00000614521.2 ENST00000424189.6 ENST00000446122.5 |
DGKI
|
diacylglycerol kinase iota |
| chr22_+_30607072 | 0.42 |
ENST00000450638.5
|
TCN2
|
transcobalamin 2 |
| chr22_+_30607167 | 0.41 |
ENST00000215838.8
|
TCN2
|
transcobalamin 2 |
| chr1_-_162023632 | 0.41 |
ENST00000367940.2
|
OLFML2B
|
olfactomedin like 2B |
| chr13_-_33285682 | 0.40 |
ENST00000336934.10
|
STARD13
|
StAR related lipid transfer domain containing 13 |
| chr4_-_88697810 | 0.40 |
ENST00000323061.7
|
NAP1L5
|
nucleosome assembly protein 1 like 5 |
| chr6_+_155149109 | 0.40 |
ENST00000456877.6
ENST00000528391.6 |
TIAM2
|
TIAM Rac1 associated GEF 2 |
| chr1_+_222928415 | 0.39 |
ENST00000284476.7
|
DISP1
|
dispatched RND transporter family member 1 |
| chr9_-_101594995 | 0.39 |
ENST00000636434.1
|
PPP3R2
|
protein phosphatase 3 regulatory subunit B, beta |
| chr22_+_35381086 | 0.39 |
ENST00000216117.9
ENST00000677931.1 ENST00000679074.1 |
HMOX1
|
heme oxygenase 1 |
| chr6_-_24910695 | 0.38 |
ENST00000643623.1
ENST00000538035.6 ENST00000647136.1 |
RIPOR2
|
RHO family interacting cell polarization regulator 2 |
| chr9_-_86947496 | 0.38 |
ENST00000298743.9
|
GAS1
|
growth arrest specific 1 |
| chr1_-_41918666 | 0.36 |
ENST00000372584.5
|
HIVEP3
|
HIVEP zinc finger 3 |
| chr4_+_85778681 | 0.36 |
ENST00000395183.6
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr2_+_30147516 | 0.36 |
ENST00000402708.5
|
YPEL5
|
yippee like 5 |
| chr12_-_55842950 | 0.36 |
ENST00000548629.5
|
MMP19
|
matrix metallopeptidase 19 |
| chrX_+_9912434 | 0.36 |
ENST00000418909.6
|
SHROOM2
|
shroom family member 2 |
| chr2_-_175005357 | 0.35 |
ENST00000409156.7
ENST00000444573.2 ENST00000409900.9 |
CHN1
|
chimerin 1 |
| chr2_-_127643212 | 0.35 |
ENST00000409286.5
|
LIMS2
|
LIM zinc finger domain containing 2 |
| chr20_+_64066211 | 0.35 |
ENST00000458442.1
|
TCEA2
|
transcription elongation factor A2 |
| chr5_-_173328407 | 0.35 |
ENST00000265087.9
|
STC2
|
stanniocalcin 2 |
| chr1_-_41918858 | 0.34 |
ENST00000372583.6
|
HIVEP3
|
HIVEP zinc finger 3 |
| chr10_-_62816309 | 0.34 |
ENST00000411732.3
|
EGR2
|
early growth response 2 |
| chr22_+_30607203 | 0.34 |
ENST00000407817.3
|
TCN2
|
transcobalamin 2 |
| chr10_-_62816341 | 0.33 |
ENST00000242480.4
ENST00000637191.1 |
EGR2
|
early growth response 2 |
| chr14_+_24115299 | 0.33 |
ENST00000559354.5
ENST00000560459.5 ENST00000559593.5 ENST00000396941.8 ENST00000396936.5 |
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr1_-_85578345 | 0.33 |
ENST00000426972.8
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr12_-_108339300 | 0.32 |
ENST00000550402.6
ENST00000552995.5 ENST00000312143.11 |
CMKLR1
|
chemerin chemokine-like receptor 1 |
| chrX_-_107775740 | 0.31 |
ENST00000372383.9
|
TSC22D3
|
TSC22 domain family member 3 |
| chr7_-_28958321 | 0.31 |
ENST00000539664.3
|
TRIL
|
TLR4 interactor with leucine rich repeats |
| chr1_-_16978276 | 0.31 |
ENST00000375534.7
|
MFAP2
|
microfibril associated protein 2 |
| chr11_+_33039996 | 0.30 |
ENST00000432887.5
ENST00000528898.1 ENST00000531632.6 |
TCP11L1
|
t-complex 11 like 1 |
| chr16_-_31094549 | 0.29 |
ENST00000394971.7
|
VKORC1
|
vitamin K epoxide reductase complex subunit 1 |
| chr2_+_148021083 | 0.28 |
ENST00000642680.2
|
MBD5
|
methyl-CpG binding domain protein 5 |
| chr13_+_31945826 | 0.28 |
ENST00000647500.1
|
FRY
|
FRY microtubule binding protein |
| chr16_-_31094727 | 0.28 |
ENST00000300851.10
ENST00000394975.3 |
VKORC1
|
vitamin K epoxide reductase complex subunit 1 |
| chr2_+_148021001 | 0.27 |
ENST00000407073.5
|
MBD5
|
methyl-CpG binding domain protein 5 |
| chr17_+_60677822 | 0.27 |
ENST00000407086.8
ENST00000589222.5 ENST00000626960.2 ENST00000390652.9 |
BCAS3
|
BCAS3 microtubule associated cell migration factor |
| chr16_-_31094890 | 0.27 |
ENST00000532364.1
ENST00000529564.1 ENST00000319788.11 ENST00000354895.4 |
ENSG00000255439.6
VKORC1
|
novel protein, VKORC1 and PRSS53 readthrough vitamin K epoxide reductase complex subunit 1 |
| chr2_+_94871419 | 0.27 |
ENST00000295201.5
|
TEKT4
|
tektin 4 |
| chr2_-_224982420 | 0.27 |
ENST00000645028.1
|
DOCK10
|
dedicator of cytokinesis 10 |
| chrX_-_102142461 | 0.26 |
ENST00000372774.7
|
TCEAL6
|
transcription elongation factor A like 6 |
| chr20_+_36091409 | 0.25 |
ENST00000202028.9
|
EPB41L1
|
erythrocyte membrane protein band 4.1 like 1 |
| chr9_+_128411715 | 0.25 |
ENST00000420034.5
ENST00000372842.5 |
CERCAM
|
cerebral endothelial cell adhesion molecule |
| chr19_-_45782388 | 0.25 |
ENST00000458663.6
|
DMPK
|
DM1 protein kinase |
| chr1_+_159780930 | 0.25 |
ENST00000368109.5
ENST00000368108.7 ENST00000368107.2 |
DUSP23
|
dual specificity phosphatase 23 |
| chr2_+_33476640 | 0.25 |
ENST00000425210.5
ENST00000444784.5 ENST00000423159.5 ENST00000403687.8 |
RASGRP3
|
RAS guanyl releasing protein 3 |
| chr11_+_45896541 | 0.25 |
ENST00000395629.2
|
MAPK8IP1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr6_-_24911029 | 0.24 |
ENST00000259698.9
ENST00000644621.1 ENST00000644411.1 |
RIPOR2
|
RHO family interacting cell polarization regulator 2 |
| chr14_-_73027117 | 0.24 |
ENST00000318876.9
|
ZFYVE1
|
zinc finger FYVE-type containing 1 |
| chr14_-_73027077 | 0.24 |
ENST00000553891.5
ENST00000556143.6 |
ZFYVE1
|
zinc finger FYVE-type containing 1 |
| chr12_+_104064520 | 0.24 |
ENST00000229330.9
|
HCFC2
|
host cell factor C2 |
| chr14_-_75981986 | 0.24 |
ENST00000238682.8
|
TGFB3
|
transforming growth factor beta 3 |
| chrX_-_150898592 | 0.24 |
ENST00000355149.8
ENST00000466436.5 ENST00000370377.8 |
CD99L2
|
CD99 molecule like 2 |
| chr16_-_21278282 | 0.23 |
ENST00000572914.2
|
CRYM
|
crystallin mu |
| chr11_+_33039561 | 0.23 |
ENST00000334274.9
|
TCP11L1
|
t-complex 11 like 1 |
| chr8_+_76681208 | 0.23 |
ENST00000651372.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr20_-_32536383 | 0.23 |
ENST00000375678.7
|
NOL4L
|
nucleolar protein 4 like |
| chr5_+_150190035 | 0.22 |
ENST00000230671.7
ENST00000524041.1 |
SLC6A7
|
solute carrier family 6 member 7 |
| chr8_-_70607654 | 0.22 |
ENST00000521425.5
|
TRAM1
|
translocation associated membrane protein 1 |
| chr11_+_6390439 | 0.22 |
ENST00000530395.1
ENST00000342245.9 ENST00000527275.5 |
SMPD1
|
sphingomyelin phosphodiesterase 1 |
| chr9_+_116153783 | 0.22 |
ENST00000328252.4
|
PAPPA
|
pappalysin 1 |
| chr2_+_56183973 | 0.22 |
ENST00000407595.3
|
CCDC85A
|
coiled-coil domain containing 85A |
| chr10_+_13100075 | 0.22 |
ENST00000378747.8
ENST00000378757.6 ENST00000378752.7 ENST00000378748.7 |
OPTN
|
optineurin |
| chr13_+_75788838 | 0.22 |
ENST00000497947.6
|
LMO7
|
LIM domain 7 |
| chr19_+_35030626 | 0.22 |
ENST00000638536.1
|
SCN1B
|
sodium voltage-gated channel beta subunit 1 |
| chr22_+_35299800 | 0.21 |
ENST00000456128.5
ENST00000411850.5 ENST00000425375.5 ENST00000449058.7 |
TOM1
|
target of myb1 membrane trafficking protein |
| chr11_+_72227881 | 0.21 |
ENST00000538751.5
ENST00000541756.5 |
INPPL1
|
inositol polyphosphate phosphatase like 1 |
| chr2_+_53971072 | 0.21 |
ENST00000422521.2
ENST00000607452.6 |
ACYP2
|
acylphosphatase 2 |
| chr3_-_64225436 | 0.21 |
ENST00000638394.2
|
PRICKLE2
|
prickle planar cell polarity protein 2 |
| chr20_+_44531758 | 0.21 |
ENST00000372891.7
ENST00000372892.7 ENST00000372894.7 |
PKIG
|
cAMP-dependent protein kinase inhibitor gamma |
| chr10_+_94089067 | 0.20 |
ENST00000371375.1
ENST00000675218.1 |
PLCE1
|
phospholipase C epsilon 1 |
| chr8_-_73878816 | 0.20 |
ENST00000602593.6
ENST00000651945.1 ENST00000419880.7 ENST00000517608.5 ENST00000650817.1 |
UBE2W
|
ubiquitin conjugating enzyme E2 W |
| chr17_-_21253398 | 0.20 |
ENST00000611551.1
|
NATD1
|
N-acetyltransferase domain containing 1 |
| chr17_+_41226648 | 0.20 |
ENST00000377721.3
|
KRTAP9-2
|
keratin associated protein 9-2 |
| chr4_+_113292838 | 0.20 |
ENST00000672411.1
ENST00000673231.1 |
ANK2
|
ankyrin 2 |
| chr1_-_150876571 | 0.20 |
ENST00000354396.6
ENST00000358595.10 ENST00000505755.5 |
ARNT
|
aryl hydrocarbon receptor nuclear translocator |
| chr10_+_94089034 | 0.20 |
ENST00000676102.1
ENST00000371385.8 |
PLCE1
|
phospholipase C epsilon 1 |
| chr8_-_92017637 | 0.20 |
ENST00000422361.6
|
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1 |
| chr19_+_35031263 | 0.19 |
ENST00000640135.1
ENST00000596348.2 |
SCN1B
|
sodium voltage-gated channel beta subunit 1 |
| chr8_-_134510182 | 0.19 |
ENST00000521673.5
|
ZFAT
|
zinc finger and AT-hook domain containing |
| chr4_+_113292925 | 0.19 |
ENST00000673353.1
ENST00000505342.6 ENST00000672915.1 ENST00000509550.5 |
ANK2
|
ankyrin 2 |
| chr10_-_90921079 | 0.19 |
ENST00000371697.4
|
ANKRD1
|
ankyrin repeat domain 1 |
| chr4_+_37891060 | 0.18 |
ENST00000261439.9
ENST00000508802.5 ENST00000402522.1 |
TBC1D1
|
TBC1 domain family member 1 |
| chr14_+_52553273 | 0.18 |
ENST00000542169.6
ENST00000555622.1 |
GPR137C
|
G protein-coupled receptor 137C |
| chr1_-_150876697 | 0.18 |
ENST00000515192.5
|
ARNT
|
aryl hydrocarbon receptor nuclear translocator |
| chr6_-_110415539 | 0.18 |
ENST00000368923.8
ENST00000368924.9 |
DDO
|
D-aspartate oxidase |
| chr15_+_92904447 | 0.18 |
ENST00000626782.2
|
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr12_+_68808143 | 0.18 |
ENST00000258149.11
ENST00000428863.6 ENST00000393412.7 |
MDM2
|
MDM2 proto-oncogene |
| chr12_-_49904217 | 0.18 |
ENST00000550635.6
|
FAIM2
|
Fas apoptotic inhibitory molecule 2 |
| chr12_-_55728977 | 0.18 |
ENST00000552164.5
|
CD63
|
CD63 molecule |
| chr14_+_22598224 | 0.18 |
ENST00000428304.6
ENST00000542041.1 ENST00000216327.10 |
ABHD4
|
abhydrolase domain containing 4, N-acyl phospholipase B |
| chr12_+_12785652 | 0.18 |
ENST00000356591.5
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr14_+_52552830 | 0.17 |
ENST00000321662.11
|
GPR137C
|
G protein-coupled receptor 137C |
| chr17_+_44170695 | 0.17 |
ENST00000293414.6
|
ASB16
|
ankyrin repeat and SOCS box containing 16 |
| chr9_+_121268060 | 0.17 |
ENST00000373808.8
ENST00000432226.7 ENST00000449733.7 |
GSN
|
gelsolin |
| chr1_-_150697128 | 0.17 |
ENST00000427665.1
ENST00000271732.8 |
GOLPH3L
|
golgi phosphoprotein 3 like |
| chr3_+_191329342 | 0.17 |
ENST00000392455.9
|
CCDC50
|
coiled-coil domain containing 50 |
| chr19_-_45782479 | 0.17 |
ENST00000447742.6
ENST00000354227.9 ENST00000291270.9 ENST00000683086.1 ENST00000343373.10 |
DMPK
|
DM1 protein kinase |
| chr17_+_47209035 | 0.17 |
ENST00000572316.5
ENST00000354968.5 ENST00000576874.5 ENST00000536623.6 |
MYL4
|
myosin light chain 4 |
| chr20_-_18497218 | 0.17 |
ENST00000337227.9
|
RBBP9
|
RB binding protein 9, serine hydrolase |
| chr6_+_69232406 | 0.17 |
ENST00000238918.12
|
ADGRB3
|
adhesion G protein-coupled receptor B3 |
| chrX_+_136197039 | 0.17 |
ENST00000370683.6
|
FHL1
|
four and a half LIM domains 1 |
| chr10_+_74824919 | 0.17 |
ENST00000648828.1
ENST00000648725.1 |
KAT6B
|
lysine acetyltransferase 6B |
| chr11_-_67353503 | 0.16 |
ENST00000539074.1
ENST00000530584.5 ENST00000531239.2 ENST00000312419.8 ENST00000529704.5 |
POLD4
|
DNA polymerase delta 4, accessory subunit |
| chrX_+_136196750 | 0.16 |
ENST00000539015.5
|
FHL1
|
four and a half LIM domains 1 |
| chr10_+_1056776 | 0.16 |
ENST00000650072.1
|
WDR37
|
WD repeat domain 37 |
| chrX_+_136197020 | 0.16 |
ENST00000370676.7
|
FHL1
|
four and a half LIM domains 1 |
| chr4_+_61200318 | 0.16 |
ENST00000683033.1
|
ADGRL3
|
adhesion G protein-coupled receptor L3 |
| chr1_-_19312068 | 0.16 |
ENST00000330072.9
ENST00000235835.8 |
AKR7A2
|
aldo-keto reductase family 7 member A2 |
| chr12_-_76486061 | 0.16 |
ENST00000548341.5
|
OSBPL8
|
oxysterol binding protein like 8 |
| chr16_-_79600698 | 0.16 |
ENST00000393350.1
|
MAF
|
MAF bZIP transcription factor |
| chr2_+_85753984 | 0.16 |
ENST00000306279.4
|
ATOH8
|
atonal bHLH transcription factor 8 |
| chr12_+_26121984 | 0.16 |
ENST00000538142.5
|
SSPN
|
sarcospan |
| chr14_-_52552493 | 0.16 |
ENST00000281741.9
ENST00000557374.1 |
TXNDC16
|
thioredoxin domain containing 16 |
| chr9_+_2159850 | 0.16 |
ENST00000416751.2
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr7_-_29990113 | 0.16 |
ENST00000426154.5
ENST00000421434.5 ENST00000434476.6 |
SCRN1
|
secernin 1 |
| chr10_+_22316445 | 0.16 |
ENST00000448361.5
|
COMMD3
|
COMM domain containing 3 |
| chr12_-_55728994 | 0.16 |
ENST00000257857.9
|
CD63
|
CD63 molecule |
| chr19_+_18588789 | 0.16 |
ENST00000450195.6
ENST00000358607.11 |
REX1BD
|
required for excision 1-B domain containing |
| chr12_+_32502114 | 0.16 |
ENST00000682739.1
ENST00000427716.7 ENST00000583694.2 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr17_-_44268119 | 0.16 |
ENST00000399246.3
ENST00000262418.12 |
SLC4A1
|
solute carrier family 4 member 1 (Diego blood group) |
| chr9_+_89605004 | 0.16 |
ENST00000252506.11
ENST00000375769.1 |
GADD45G
|
growth arrest and DNA damage inducible gamma |
| chr17_+_6444441 | 0.16 |
ENST00000250056.12
ENST00000571373.5 ENST00000570337.6 ENST00000572595.6 ENST00000572447.6 ENST00000576056.5 |
PIMREG
|
PICALM interacting mitotic regulator |
| chr18_+_3448456 | 0.16 |
ENST00000549780.5
|
TGIF1
|
TGFB induced factor homeobox 1 |
| chr16_+_30372291 | 0.15 |
ENST00000568749.5
|
MYLPF
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr7_+_18496231 | 0.15 |
ENST00000401921.5
|
HDAC9
|
histone deacetylase 9 |
| chr1_-_243163310 | 0.15 |
ENST00000492145.1
ENST00000490813.5 ENST00000464936.5 |
CEP170
|
centrosomal protein 170 |
| chr11_-_1486763 | 0.15 |
ENST00000329957.7
|
MOB2
|
MOB kinase activator 2 |
| chr3_-_100114488 | 0.15 |
ENST00000477258.2
ENST00000354552.7 ENST00000331335.9 ENST00000398326.2 |
FILIP1L
|
filamin A interacting protein 1 like |
| chr2_+_61905646 | 0.15 |
ENST00000311832.5
|
COMMD1
|
copper metabolism domain containing 1 |
| chr2_-_65366650 | 0.15 |
ENST00000443619.6
|
SPRED2
|
sprouty related EVH1 domain containing 2 |
| chr9_-_101594918 | 0.15 |
ENST00000374806.2
|
PPP3R2
|
protein phosphatase 3 regulatory subunit B, beta |
| chr15_+_75336018 | 0.15 |
ENST00000567195.5
|
COMMD4
|
COMM domain containing 4 |
| chr14_+_102922639 | 0.15 |
ENST00000299155.10
|
AMN
|
amnion associated transmembrane protein |
| chr7_-_76626127 | 0.14 |
ENST00000454397.1
|
POMZP3
|
POM121 and ZP3 fusion |
| chr22_+_30694851 | 0.14 |
ENST00000332585.11
|
OSBP2
|
oxysterol binding protein 2 |
| chr11_-_62556230 | 0.14 |
ENST00000530285.5
|
AHNAK
|
AHNAK nucleoprotein |
| chr17_-_81937221 | 0.14 |
ENST00000402252.6
ENST00000583564.5 ENST00000585244.1 ENST00000337943.9 ENST00000579698.5 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chr10_+_22316375 | 0.14 |
ENST00000376836.8
ENST00000456711.5 ENST00000444869.5 ENST00000475460.6 ENST00000602390.5 ENST00000489125.2 |
COMMD3
COMMD3-BMI1
|
COMM domain containing 3 COMMD3-BMI1 readthrough |
| chr5_+_50666917 | 0.14 |
ENST00000514342.6
|
PARP8
|
poly(ADP-ribose) polymerase family member 8 |
| chr5_+_138084015 | 0.14 |
ENST00000504809.5
ENST00000506684.6 ENST00000398754.1 |
WNT8A
|
Wnt family member 8A |
| chr5_+_141969074 | 0.14 |
ENST00000506938.5
ENST00000394520.7 ENST00000394514.6 ENST00000512565.5 |
RNF14
|
ring finger protein 14 |
| chr3_+_54123452 | 0.14 |
ENST00000620722.4
ENST00000490478.5 |
CACNA2D3
|
calcium voltage-gated channel auxiliary subunit alpha2delta 3 |
| chr2_-_27071628 | 0.14 |
ENST00000447619.5
ENST00000429985.1 ENST00000456793.2 |
OST4
|
oligosaccharyltransferase complex subunit 4, non-catalytic |
| chr4_-_52038260 | 0.14 |
ENST00000381431.10
|
SGCB
|
sarcoglycan beta |
| chr6_+_52423680 | 0.14 |
ENST00000538167.2
|
EFHC1
|
EF-hand domain containing 1 |
| chr7_-_29989774 | 0.14 |
ENST00000242059.10
|
SCRN1
|
secernin 1 |
| chr15_-_31870651 | 0.14 |
ENST00000307050.6
ENST00000560598.2 |
OTUD7A
|
OTU deubiquitinase 7A |
| chr7_-_30550761 | 0.14 |
ENST00000598361.4
ENST00000440438.6 |
ENSG00000281039.1
GARS1-DT
|
novel protein GARS1 divergent transcript |
| chr5_-_88785493 | 0.14 |
ENST00000503554.4
|
MEF2C
|
myocyte enhancer factor 2C |
| chr15_+_75336078 | 0.14 |
ENST00000567377.5
ENST00000562789.5 ENST00000568301.1 |
COMMD4
|
COMM domain containing 4 |
| chr9_+_2159672 | 0.14 |
ENST00000634343.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr6_+_46793379 | 0.14 |
ENST00000230588.9
ENST00000611727.2 |
MEP1A
|
meprin A subunit alpha |
| chr1_+_174875505 | 0.14 |
ENST00000486220.5
|
RABGAP1L
|
RAB GTPase activating protein 1 like |
| chr16_-_79600727 | 0.13 |
ENST00000326043.5
|
MAF
|
MAF bZIP transcription factor |
| chr17_-_81937320 | 0.13 |
ENST00000577624.5
ENST00000403172.8 ENST00000619204.4 ENST00000629768.2 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chrX_+_65588368 | 0.13 |
ENST00000609672.5
|
MSN
|
moesin |
| chr17_+_35243063 | 0.13 |
ENST00000542451.1
ENST00000299977.9 ENST00000592325.1 |
SLFN5
|
schlafen family member 5 |
| chr19_-_33521749 | 0.13 |
ENST00000588328.6
ENST00000651901.1 ENST00000651646.1 ENST00000436370.7 ENST00000244137.12 ENST00000397032.8 |
PEPD
|
peptidase D |
| chr4_-_139302516 | 0.13 |
ENST00000394228.5
ENST00000539387.5 |
NDUFC1
|
NADH:ubiquinone oxidoreductase subunit C1 |
| chr8_+_80486209 | 0.13 |
ENST00000426744.5
ENST00000455036.8 |
ZBTB10
|
zinc finger and BTB domain containing 10 |
| chr17_-_81937277 | 0.13 |
ENST00000405481.8
ENST00000329875.13 ENST00000585215.5 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chr15_+_75336053 | 0.13 |
ENST00000564815.5
ENST00000338995.10 ENST00000267935.13 |
COMMD4
|
COMM domain containing 4 |
| chr9_-_90642855 | 0.13 |
ENST00000637905.1
|
DIRAS2
|
DIRAS family GTPase 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TFAP4_MSC
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:1904761 | negative regulation of myofibroblast differentiation(GO:1904761) |
| 0.2 | 0.7 | GO:0021593 | rhombomere morphogenesis(GO:0021593) rhombomere 3 morphogenesis(GO:0021658) |
| 0.2 | 0.4 | GO:0014806 | smooth muscle hyperplasia(GO:0014806) |
| 0.2 | 1.1 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.2 | 3.3 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 0.9 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.1 | 1.6 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.1 | 0.4 | GO:0035638 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.1 | 0.6 | GO:2001107 | negative regulation of establishment of T cell polarity(GO:1903904) negative regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001107) |
| 0.1 | 1.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 1.7 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.1 | 1.0 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 0.9 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 0.2 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.1 | 0.2 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.1 | 0.2 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.1 | 0.5 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.1 | 0.3 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.1 | 0.1 | GO:0061317 | canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) |
| 0.1 | 0.5 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.1 | 0.4 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 0.8 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.1 | 1.0 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.2 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.1 | 0.4 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 1.0 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.2 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.2 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.4 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.6 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.2 | GO:0036369 | transcription factor catabolic process(GO:0036369) cellular response to actinomycin D(GO:0072717) |
| 0.0 | 0.4 | GO:0001554 | luteolysis(GO:0001554) |
| 0.0 | 0.3 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.0 | 0.4 | GO:0046886 | positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.0 | 0.1 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.4 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.2 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.0 | 0.6 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.0 | GO:1902954 | regulation of early endosome to recycling endosome transport(GO:1902954) |
| 0.0 | 0.2 | GO:1903906 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.1 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:1902966 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.0 | 0.5 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.2 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.0 | 1.4 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 0.2 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.1 | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 0.0 | 0.1 | GO:0099403 | negative regulation of maintenance of sister chromatid cohesion(GO:0034092) negative regulation of maintenance of mitotic sister chromatid cohesion(GO:0034183) maintenance of mitotic sister chromatid cohesion, telomeric(GO:0099403) mitotic sister chromatid cohesion, telomeric(GO:0099404) regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904907) negative regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904908) |
| 0.0 | 0.1 | GO:0090156 | cellular sphingolipid homeostasis(GO:0090156) |
| 0.0 | 0.2 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.4 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.1 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.4 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.4 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.1 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.7 | GO:2001046 | positive regulation of integrin-mediated signaling pathway(GO:2001046) |
| 0.0 | 0.1 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.1 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) positive regulation of B cell apoptotic process(GO:0002904) regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.1 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.1 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) vagus nerve development(GO:0021564) |
| 0.0 | 0.2 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.1 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.2 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 | 0.1 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.0 | GO:0060913 | visceral motor neuron differentiation(GO:0021524) cardiac cell fate determination(GO:0060913) positive regulation of granulocyte colony-stimulating factor production(GO:0071657) positive regulation of macrophage colony-stimulating factor production(GO:1901258) |
| 0.0 | 0.2 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.1 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.2 | GO:0030638 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 0.6 | GO:0046596 | regulation of viral entry into host cell(GO:0046596) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.3 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.2 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.3 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 | 0.1 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.1 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 0.1 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.1 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.0 | GO:0002905 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.3 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.3 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.1 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.1 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.0 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.1 | GO:0071964 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.0 | GO:0016334 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 0.0 | 0.0 | GO:0090274 | regulation of somatostatin secretion(GO:0090273) positive regulation of somatostatin secretion(GO:0090274) |
| 0.0 | 0.0 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.1 | GO:0009838 | abscission(GO:0009838) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0034959 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 0.1 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.3 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.2 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 1.0 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 0.9 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.6 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 1.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.7 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.2 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.3 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.6 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.2 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.1 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.0 | 0.7 | GO:0097431 | pericentriolar material(GO:0000242) mitotic spindle pole(GO:0097431) |
| 0.0 | 1.2 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.3 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 2.0 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.0 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 2.3 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.1 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.2 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:0072357 | glycogen granule(GO:0042587) PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.1 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.3 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 0.4 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.1 | 0.8 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.1 | 1.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 1.6 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 1.0 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.5 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.1 | 0.3 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.1 | 0.4 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.2 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.2 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.1 | 0.4 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.3 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.0 | 0.2 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.2 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.9 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 1.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0052815 | medium-chain acyl-CoA hydrolase activity(GO:0052815) long-chain acyl-CoA hydrolase activity(GO:0052816) |
| 0.0 | 0.1 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.0 | 0.2 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.1 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.0 | 0.2 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.6 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.4 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.0 | 0.6 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.7 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.4 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.5 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.4 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.0 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.0 | 0.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.0 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.5 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.0 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.3 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.0 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.7 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.6 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |