Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for UGAAAUG
Z-value: 0.66
miRNA associated with seed UGAAAUG
| Name | miRBASE accession |
|---|---|
|
hsa-miR-203a-3p.2
|
Activity profile of UGAAAUG motif
Sorted Z-values of UGAAAUG motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_72282457 | 0.52 |
ENST00000357731.10
|
NEGR1
|
neuronal growth regulator 1 |
| chr5_-_9546066 | 0.47 |
ENST00000382496.10
ENST00000652226.1 |
SEMA5A
|
semaphorin 5A |
| chr1_-_56579555 | 0.36 |
ENST00000371250.4
|
PLPP3
|
phospholipid phosphatase 3 |
| chr21_-_37916440 | 0.31 |
ENST00000609713.2
|
KCNJ6
|
potassium inwardly rectifying channel subfamily J member 6 |
| chr5_+_83471736 | 0.26 |
ENST00000265077.8
|
VCAN
|
versican |
| chr2_-_224947030 | 0.20 |
ENST00000409592.7
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr8_+_17027230 | 0.17 |
ENST00000318063.10
|
MICU3
|
mitochondrial calcium uptake family member 3 |
| chr5_-_169980474 | 0.16 |
ENST00000377365.4
|
INSYN2B
|
inhibitory synaptic factor family member 2B |
| chr4_+_122826679 | 0.16 |
ENST00000264498.8
|
FGF2
|
fibroblast growth factor 2 |
| chr6_-_139374605 | 0.16 |
ENST00000618718.1
ENST00000367651.4 |
CITED2
|
Cbp/p300 interacting transactivator with Glu/Asp rich carboxy-terminal domain 2 |
| chr5_-_122078249 | 0.15 |
ENST00000231004.5
|
LOX
|
lysyl oxidase |
| chr17_+_48997377 | 0.15 |
ENST00000290341.8
|
IGF2BP1
|
insulin like growth factor 2 mRNA binding protein 1 |
| chr2_-_40452046 | 0.14 |
ENST00000406785.6
|
SLC8A1
|
solute carrier family 8 member A1 |
| chr5_-_159099909 | 0.14 |
ENST00000313708.11
|
EBF1
|
EBF transcription factor 1 |
| chr1_+_212950572 | 0.14 |
ENST00000366968.8
ENST00000490792.1 ENST00000366964.7 |
VASH2
|
vasohibin 2 |
| chr10_-_62268837 | 0.14 |
ENST00000373789.8
|
RTKN2
|
rhotekin 2 |
| chrX_-_19122559 | 0.13 |
ENST00000357544.7
ENST00000360279.8 ENST00000379873.6 ENST00000379876.5 ENST00000379878.7 ENST00000354791.7 |
ADGRG2
|
adhesion G protein-coupled receptor G2 |
| chr2_-_178478541 | 0.12 |
ENST00000424785.7
|
FKBP7
|
FKBP prolyl isomerase 7 |
| chr2_-_226799806 | 0.11 |
ENST00000305123.6
|
IRS1
|
insulin receptor substrate 1 |
| chr1_+_210232776 | 0.11 |
ENST00000367012.4
|
SERTAD4
|
SERTA domain containing 4 |
| chr21_+_38256698 | 0.11 |
ENST00000613499.4
ENST00000612702.4 ENST00000398925.5 ENST00000398928.5 ENST00000328656.8 ENST00000443341.5 |
KCNJ15
|
potassium inwardly rectifying channel subfamily J member 15 |
| chr2_+_148021001 | 0.11 |
ENST00000407073.5
|
MBD5
|
methyl-CpG binding domain protein 5 |
| chr10_+_74826550 | 0.11 |
ENST00000649657.1
ENST00000372714.6 ENST00000649442.1 ENST00000648539.1 ENST00000647666.1 ENST00000648048.1 ENST00000287239.10 ENST00000649375.1 |
KAT6B
|
lysine acetyltransferase 6B |
| chr16_+_15434577 | 0.11 |
ENST00000300006.9
|
BMERB1
|
bMERB domain containing 1 |
| chr1_+_63773966 | 0.10 |
ENST00000371079.6
ENST00000371080.5 |
ROR1
|
receptor tyrosine kinase like orphan receptor 1 |
| chr7_+_17298642 | 0.10 |
ENST00000242057.9
|
AHR
|
aryl hydrocarbon receptor |
| chr14_+_20684547 | 0.09 |
ENST00000555835.3
ENST00000397995.2 ENST00000553909.1 |
RNASE4
ENSG00000259171.1
|
ribonuclease A family member 4 novel protein, ANG-RNASE4 readthrough |
| chr7_+_7968787 | 0.09 |
ENST00000223145.10
|
GLCCI1
|
glucocorticoid induced 1 |
| chr8_+_41578176 | 0.09 |
ENST00000396987.7
ENST00000519853.5 |
GPAT4
|
glycerol-3-phosphate acyltransferase 4 |
| chr5_-_124744513 | 0.09 |
ENST00000504926.5
|
ZNF608
|
zinc finger protein 608 |
| chr5_+_149141483 | 0.09 |
ENST00000326685.11
ENST00000309868.12 |
ABLIM3
|
actin binding LIM protein family member 3 |
| chr16_-_73048104 | 0.09 |
ENST00000268489.10
|
ZFHX3
|
zinc finger homeobox 3 |
| chr5_-_59893718 | 0.09 |
ENST00000340635.11
|
PDE4D
|
phosphodiesterase 4D |
| chr7_-_13989658 | 0.09 |
ENST00000430479.6
ENST00000433547.1 ENST00000405192.6 |
ETV1
|
ETS variant transcription factor 1 |
| chr17_+_66302606 | 0.09 |
ENST00000413366.8
|
PRKCA
|
protein kinase C alpha |
| chr12_+_32502114 | 0.08 |
ENST00000682739.1
ENST00000427716.7 ENST00000583694.2 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr17_-_55722857 | 0.08 |
ENST00000424486.3
|
TMEM100
|
transmembrane protein 100 |
| chr11_+_108223112 | 0.08 |
ENST00000452508.6
ENST00000683914.1 ENST00000683150.1 ENST00000639953.1 ENST00000640388.1 ENST00000639240.1 |
ATM
|
ATM serine/threonine kinase |
| chr5_-_39424966 | 0.08 |
ENST00000515700.5
ENST00000320816.11 ENST00000339788.10 |
DAB2
|
DAB adaptor protein 2 |
| chrX_-_84502442 | 0.08 |
ENST00000297977.9
ENST00000506585.6 ENST00000373177.3 ENST00000449553.2 |
HDX
|
highly divergent homeobox |
| chr5_-_151686908 | 0.08 |
ENST00000231061.9
|
SPARC
|
secreted protein acidic and cysteine rich |
| chr2_-_179264757 | 0.08 |
ENST00000428443.8
|
SESTD1
|
SEC14 and spectrin domain containing 1 |
| chr5_-_65722094 | 0.08 |
ENST00000381007.9
|
SGTB
|
small glutamine rich tetratricopeptide repeat containing beta |
| chr16_-_17470953 | 0.08 |
ENST00000261381.7
|
XYLT1
|
xylosyltransferase 1 |
| chr17_-_35089212 | 0.08 |
ENST00000584655.5
ENST00000447669.6 ENST00000315249.11 |
RFFL
|
ring finger and FYVE like domain containing E3 ubiquitin protein ligase |
| chr18_-_28177102 | 0.07 |
ENST00000413878.2
ENST00000269141.8 |
CDH2
|
cadherin 2 |
| chr12_+_119668109 | 0.07 |
ENST00000229328.10
ENST00000630317.1 |
PRKAB1
|
protein kinase AMP-activated non-catalytic subunit beta 1 |
| chr1_+_25430854 | 0.07 |
ENST00000399766.7
|
MACO1
|
macoilin 1 |
| chr4_-_184826030 | 0.07 |
ENST00000507295.5
ENST00000504900.5 ENST00000454703.6 |
ACSL1
|
acyl-CoA synthetase long chain family member 1 |
| chr17_+_67825664 | 0.07 |
ENST00000321892.8
|
BPTF
|
bromodomain PHD finger transcription factor |
| chr14_-_52552493 | 0.07 |
ENST00000281741.9
ENST00000557374.1 |
TXNDC16
|
thioredoxin domain containing 16 |
| chr14_+_57268963 | 0.07 |
ENST00000261558.8
|
AP5M1
|
adaptor related protein complex 5 subunit mu 1 |
| chr16_+_53054973 | 0.07 |
ENST00000447540.6
ENST00000615216.4 ENST00000566029.5 |
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr3_+_191329342 | 0.07 |
ENST00000392455.9
|
CCDC50
|
coiled-coil domain containing 50 |
| chr5_+_173888335 | 0.07 |
ENST00000265085.10
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr8_-_115668966 | 0.07 |
ENST00000395715.8
|
TRPS1
|
transcriptional repressor GATA binding 1 |
| chr13_-_67230377 | 0.07 |
ENST00000544246.5
ENST00000377861.4 |
PCDH9
|
protocadherin 9 |
| chr14_+_85530127 | 0.07 |
ENST00000330753.6
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr20_-_49482645 | 0.07 |
ENST00000371741.6
|
KCNB1
|
potassium voltage-gated channel subfamily B member 1 |
| chr5_-_140557414 | 0.06 |
ENST00000336283.9
|
SRA1
|
steroid receptor RNA activator 1 |
| chr3_-_115071333 | 0.06 |
ENST00000462705.5
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr8_-_92103217 | 0.06 |
ENST00000615601.4
ENST00000523629.5 |
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1 |
| chr14_+_105314711 | 0.06 |
ENST00000447393.6
ENST00000547217.5 |
PACS2
|
phosphofurin acidic cluster sorting protein 2 |
| chr6_-_57221402 | 0.06 |
ENST00000317483.4
|
RAB23
|
RAB23, member RAS oncogene family |
| chr6_-_52577012 | 0.06 |
ENST00000182527.4
|
TRAM2
|
translocation associated membrane protein 2 |
| chr1_+_62437015 | 0.06 |
ENST00000339950.5
|
USP1
|
ubiquitin specific peptidase 1 |
| chr13_+_34942263 | 0.06 |
ENST00000379939.7
ENST00000400445.7 |
NBEA
|
neurobeachin |
| chr5_-_132963621 | 0.06 |
ENST00000265343.10
|
AFF4
|
AF4/FMR2 family member 4 |
| chr4_+_141636563 | 0.06 |
ENST00000320650.9
ENST00000296545.11 |
IL15
|
interleukin 15 |
| chr15_+_76336755 | 0.06 |
ENST00000290759.9
|
ISL2
|
ISL LIM homeobox 2 |
| chr1_+_100896060 | 0.06 |
ENST00000370112.8
ENST00000357650.9 |
SLC30A7
|
solute carrier family 30 member 7 |
| chr3_+_139935176 | 0.06 |
ENST00000458420.7
|
CLSTN2
|
calsyntenin 2 |
| chr1_+_218345326 | 0.06 |
ENST00000366930.9
|
TGFB2
|
transforming growth factor beta 2 |
| chr10_+_1049476 | 0.06 |
ENST00000358220.5
|
WDR37
|
WD repeat domain 37 |
| chr11_-_58844695 | 0.05 |
ENST00000287275.6
|
GLYATL2
|
glycine-N-acyltransferase like 2 |
| chr8_+_94719865 | 0.05 |
ENST00000414645.6
|
DPY19L4
|
dpy-19 like 4 |
| chr9_+_111896804 | 0.05 |
ENST00000374279.4
|
UGCG
|
UDP-glucose ceramide glucosyltransferase |
| chr10_+_103277129 | 0.05 |
ENST00000369849.9
|
INA
|
internexin neuronal intermediate filament protein alpha |
| chr1_-_182400660 | 0.05 |
ENST00000367565.2
|
TEDDM1
|
transmembrane epididymal protein 1 |
| chr10_+_123008966 | 0.05 |
ENST00000368869.8
ENST00000358776.7 |
ACADSB
|
acyl-CoA dehydrogenase short/branched chain |
| chr2_+_11746576 | 0.05 |
ENST00000256720.6
ENST00000674199.1 ENST00000441684.5 ENST00000423495.1 |
LPIN1
|
lipin 1 |
| chr2_+_148644706 | 0.05 |
ENST00000258484.11
|
EPC2
|
enhancer of polycomb homolog 2 |
| chr5_-_38595396 | 0.05 |
ENST00000263409.8
|
LIFR
|
LIF receptor subunit alpha |
| chr5_+_128083757 | 0.05 |
ENST00000262461.7
ENST00000628403.2 ENST00000343225.4 |
SLC12A2
|
solute carrier family 12 member 2 |
| chr6_-_75206044 | 0.05 |
ENST00000322507.13
|
COL12A1
|
collagen type XII alpha 1 chain |
| chr14_+_73644875 | 0.05 |
ENST00000554113.5
ENST00000553645.7 ENST00000555631.6 ENST00000311089.7 ENST00000555919.7 ENST00000554339.5 ENST00000554871.5 |
DNAL1
|
dynein axonemal light chain 1 |
| chr11_-_106022209 | 0.05 |
ENST00000301919.9
ENST00000534458.1 ENST00000530108.1 ENST00000530788.1 |
MSANTD4
|
Myb/SANT DNA binding domain containing 4 with coiled-coils |
| chr18_+_70288991 | 0.05 |
ENST00000397942.4
|
SOCS6
|
suppressor of cytokine signaling 6 |
| chrX_-_78139612 | 0.05 |
ENST00000341864.6
|
TAF9B
|
TATA-box binding protein associated factor 9b |
| chr2_-_36966503 | 0.05 |
ENST00000263918.9
|
STRN
|
striatin |
| chr3_+_45594727 | 0.05 |
ENST00000273317.5
|
LIMD1
|
LIM domains containing 1 |
| chr3_-_64445396 | 0.05 |
ENST00000295902.11
|
PRICKLE2
|
prickle planar cell polarity protein 2 |
| chr3_+_72888031 | 0.05 |
ENST00000389617.9
|
GXYLT2
|
glucoside xylosyltransferase 2 |
| chr12_+_104064520 | 0.04 |
ENST00000229330.9
|
HCFC2
|
host cell factor C2 |
| chr17_-_78360066 | 0.04 |
ENST00000587578.1
ENST00000330871.3 |
SOCS3
|
suppressor of cytokine signaling 3 |
| chr14_-_73714361 | 0.04 |
ENST00000316836.5
|
PNMA1
|
PNMA family member 1 |
| chr12_-_109573482 | 0.04 |
ENST00000540016.5
ENST00000545712.7 |
MMAB
|
metabolism of cobalamin associated B |
| chr20_+_41028814 | 0.04 |
ENST00000361337.3
|
TOP1
|
DNA topoisomerase I |
| chr15_+_91100194 | 0.04 |
ENST00000394232.6
|
SV2B
|
synaptic vesicle glycoprotein 2B |
| chr4_+_38867789 | 0.04 |
ENST00000358869.5
|
FAM114A1
|
family with sequence similarity 114 member A1 |
| chr8_-_143840940 | 0.04 |
ENST00000442628.7
|
NRBP2
|
nuclear receptor binding protein 2 |
| chr6_+_73695779 | 0.04 |
ENST00000422508.6
ENST00000437994.6 |
CD109
|
CD109 molecule |
| chr11_+_130448633 | 0.04 |
ENST00000299164.4
|
ADAMTS15
|
ADAM metallopeptidase with thrombospondin type 1 motif 15 |
| chr5_-_127030545 | 0.04 |
ENST00000308660.6
|
MARCHF3
|
membrane associated ring-CH-type finger 3 |
| chr8_+_78516329 | 0.04 |
ENST00000396418.7
ENST00000352966.9 |
PKIA
|
cAMP-dependent protein kinase inhibitor alpha |
| chr5_+_144205250 | 0.04 |
ENST00000507359.3
|
KCTD16
|
potassium channel tetramerization domain containing 16 |
| chr12_-_64752871 | 0.04 |
ENST00000418919.6
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase |
| chr15_+_85380565 | 0.04 |
ENST00000559362.5
ENST00000394518.7 |
AKAP13
|
A-kinase anchoring protein 13 |
| chr6_-_84227596 | 0.04 |
ENST00000257766.8
|
CEP162
|
centrosomal protein 162 |
| chr13_-_109786567 | 0.04 |
ENST00000375856.5
|
IRS2
|
insulin receptor substrate 2 |
| chr12_-_111685922 | 0.04 |
ENST00000419234.9
|
BRAP
|
BRCA1 associated protein |
| chr12_+_120446418 | 0.04 |
ENST00000551765.6
ENST00000229384.5 |
GATC
|
glutamyl-tRNA amidotransferase subunit C |
| chr12_-_15221394 | 0.04 |
ENST00000537647.5
ENST00000256953.6 ENST00000546331.5 |
RERG
|
RAS like estrogen regulated growth inhibitor |
| chr7_+_66921217 | 0.04 |
ENST00000341567.8
ENST00000607045.5 |
TMEM248
|
transmembrane protein 248 |
| chrX_-_19970298 | 0.04 |
ENST00000379687.7
ENST00000379682.8 |
BCLAF3
|
BCLAF1 and THRAP3 family member 3 |
| chr20_+_58309704 | 0.04 |
ENST00000244040.4
|
RAB22A
|
RAB22A, member RAS oncogene family |
| chr12_-_109880527 | 0.04 |
ENST00000318348.9
|
GLTP
|
glycolipid transfer protein |
| chr6_+_57090069 | 0.04 |
ENST00000370708.8
ENST00000370702.5 |
ZNF451
|
zinc finger protein 451 |
| chr12_-_108731505 | 0.04 |
ENST00000261401.8
ENST00000552871.5 |
CORO1C
|
coronin 1C |
| chr18_+_657637 | 0.04 |
ENST00000323274.15
|
TYMS
|
thymidylate synthetase |
| chr19_+_13795434 | 0.04 |
ENST00000254323.6
|
ZSWIM4
|
zinc finger SWIM-type containing 4 |
| chr5_+_52787899 | 0.04 |
ENST00000274311.3
ENST00000282588.7 |
PELO
ITGA1
|
pelota mRNA surveillance and ribosome rescue factor integrin subunit alpha 1 |
| chr1_-_32702736 | 0.04 |
ENST00000373484.4
ENST00000409190.8 |
SYNC
|
syncoilin, intermediate filament protein |
| chr17_-_35121487 | 0.04 |
ENST00000593039.5
|
ENSG00000267618.5
|
RAD51L3-RFFL readthrough |
| chr5_-_90474765 | 0.04 |
ENST00000316610.7
|
MBLAC2
|
metallo-beta-lactamase domain containing 2 |
| chr17_-_31314040 | 0.03 |
ENST00000330927.5
|
EVI2B
|
ecotropic viral integration site 2B |
| chr17_+_32350132 | 0.03 |
ENST00000321233.10
ENST00000394673.6 ENST00000394670.9 ENST00000579634.5 ENST00000580759.5 ENST00000342555.10 ENST00000577908.5 ENST00000394679.9 ENST00000582165.1 |
ZNF207
|
zinc finger protein 207 |
| chr6_+_116877236 | 0.03 |
ENST00000332958.3
|
RFX6
|
regulatory factor X6 |
| chr9_+_35732649 | 0.03 |
ENST00000353704.3
|
CREB3
|
cAMP responsive element binding protein 3 |
| chr2_+_206443496 | 0.03 |
ENST00000264377.8
|
ADAM23
|
ADAM metallopeptidase domain 23 |
| chr11_+_111937320 | 0.03 |
ENST00000440460.7
|
DIXDC1
|
DIX domain containing 1 |
| chr10_+_19816395 | 0.03 |
ENST00000377252.5
|
PLXDC2
|
plexin domain containing 2 |
| chr19_-_40690629 | 0.03 |
ENST00000252891.8
|
NUMBL
|
NUMB like endocytic adaptor protein |
| chr17_-_5486157 | 0.03 |
ENST00000572834.5
ENST00000158771.9 ENST00000570848.5 ENST00000571971.1 |
DERL2
|
derlin 2 |
| chr17_-_63827647 | 0.03 |
ENST00000584574.5
ENST00000585145.1 ENST00000427159.7 |
FTSJ3
|
FtsJ RNA 2'-O-methyltransferase 3 |
| chr22_+_41976933 | 0.03 |
ENST00000396425.7
|
SEPTIN3
|
septin 3 |
| chr8_+_17246921 | 0.03 |
ENST00000518038.1
ENST00000324849.9 |
VPS37A
|
VPS37A subunit of ESCRT-I |
| chr11_+_122655712 | 0.03 |
ENST00000284273.6
|
UBASH3B
|
ubiquitin associated and SH3 domain containing B |
| chr11_+_34051722 | 0.03 |
ENST00000341394.9
ENST00000389645.7 |
CAPRIN1
|
cell cycle associated protein 1 |
| chr6_+_159969070 | 0.03 |
ENST00000356956.6
|
IGF2R
|
insulin like growth factor 2 receptor |
| chr14_-_74019255 | 0.03 |
ENST00000334696.11
ENST00000556242.5 |
ENTPD5
|
ectonucleoside triphosphate diphosphohydrolase 5 (inactive) |
| chr17_-_64662290 | 0.03 |
ENST00000262435.14
|
SMURF2
|
SMAD specific E3 ubiquitin protein ligase 2 |
| chr7_+_88759690 | 0.03 |
ENST00000333190.5
|
ZNF804B
|
zinc finger protein 804B |
| chr5_-_24644968 | 0.03 |
ENST00000264463.8
|
CDH10
|
cadherin 10 |
| chr17_+_29390326 | 0.03 |
ENST00000261716.8
|
TAOK1
|
TAO kinase 1 |
| chr10_+_7818497 | 0.03 |
ENST00000344293.6
|
TAF3
|
TATA-box binding protein associated factor 3 |
| chr1_+_168225950 | 0.03 |
ENST00000367829.5
ENST00000271375.7 ENST00000630869.1 |
SFT2D2
|
SFT2 domain containing 2 |
| chr1_+_203475798 | 0.03 |
ENST00000343110.3
|
PRELP
|
proline and arginine rich end leucine rich repeat protein |
| chr7_-_132576493 | 0.03 |
ENST00000321063.8
|
PLXNA4
|
plexin A4 |
| chr5_+_34929572 | 0.03 |
ENST00000382021.2
|
DNAJC21
|
DnaJ heat shock protein family (Hsp40) member C21 |
| chr11_-_26722051 | 0.03 |
ENST00000396005.8
|
SLC5A12
|
solute carrier family 5 member 12 |
| chr3_-_179071742 | 0.03 |
ENST00000311417.7
ENST00000652290.1 |
ZMAT3
|
zinc finger matrin-type 3 |
| chr9_+_15553002 | 0.03 |
ENST00000380701.8
|
CCDC171
|
coiled-coil domain containing 171 |
| chr10_+_113854610 | 0.03 |
ENST00000369301.3
|
NHLRC2
|
NHL repeat containing 2 |
| chr14_-_49852760 | 0.03 |
ENST00000555970.5
ENST00000298310.10 ENST00000554626.5 ENST00000556672.1 |
NEMF
|
nuclear export mediator factor |
| chr13_-_44576319 | 0.03 |
ENST00000458659.3
|
TSC22D1
|
TSC22 domain family member 1 |
| chr17_-_41481140 | 0.03 |
ENST00000246639.6
ENST00000393989.1 |
KRT35
|
keratin 35 |
| chr1_+_93345893 | 0.03 |
ENST00000370272.9
ENST00000370267.1 |
DR1
|
down-regulator of transcription 1 |
| chrX_+_46573757 | 0.03 |
ENST00000276055.4
|
CHST7
|
carbohydrate sulfotransferase 7 |
| chrX_-_120469282 | 0.03 |
ENST00000200639.9
ENST00000371335.4 |
LAMP2
|
lysosomal associated membrane protein 2 |
| chr17_+_59619885 | 0.03 |
ENST00000269122.8
ENST00000580081.1 ENST00000579456.5 |
CLTC
|
clathrin heavy chain |
| chr5_+_72816643 | 0.03 |
ENST00000337273.10
ENST00000523768.5 |
TNPO1
|
transportin 1 |
| chr2_-_55419565 | 0.03 |
ENST00000647341.1
ENST00000647401.1 ENST00000336838.10 ENST00000621814.4 ENST00000644033.1 ENST00000645477.1 ENST00000647517.1 |
CCDC88A
|
coiled-coil domain containing 88A |
| chr20_+_35226676 | 0.03 |
ENST00000246186.8
|
MMP24
|
matrix metallopeptidase 24 |
| chrX_-_46759055 | 0.03 |
ENST00000328306.4
ENST00000616978.5 |
SLC9A7
|
solute carrier family 9 member A7 |
| chr14_-_53152371 | 0.03 |
ENST00000323669.10
|
DDHD1
|
DDHD domain containing 1 |
| chr20_-_5610980 | 0.02 |
ENST00000379019.7
|
GPCPD1
|
glycerophosphocholine phosphodiesterase 1 |
| chr4_+_81030700 | 0.02 |
ENST00000282701.4
|
BMP3
|
bone morphogenetic protein 3 |
| chr6_-_145814744 | 0.02 |
ENST00000237281.5
|
FBXO30
|
F-box protein 30 |
| chr12_+_53454718 | 0.02 |
ENST00000552819.5
ENST00000455667.7 |
PCBP2
|
poly(rC) binding protein 2 |
| chr9_+_32384603 | 0.02 |
ENST00000541043.5
ENST00000379923.5 ENST00000309951.8 |
ACO1
|
aconitase 1 |
| chr15_-_52529050 | 0.02 |
ENST00000399231.7
|
MYO5A
|
myosin VA |
| chrX_-_15854791 | 0.02 |
ENST00000545766.7
ENST00000380291.5 ENST00000672987.1 ENST00000329235.6 |
AP1S2
|
adaptor related protein complex 1 subunit sigma 2 |
| chrX_+_74421450 | 0.02 |
ENST00000587091.6
|
SLC16A2
|
solute carrier family 16 member 2 |
| chr12_-_56258327 | 0.02 |
ENST00000267116.8
|
ANKRD52
|
ankyrin repeat domain 52 |
| chr20_+_62143729 | 0.02 |
ENST00000331758.8
ENST00000450482.5 |
SS18L1
|
SS18L1 subunit of BAF chromatin remodeling complex |
| chr12_+_121400041 | 0.02 |
ENST00000361234.9
ENST00000613529.4 |
RNF34
|
ring finger protein 34 |
| chr7_-_14903319 | 0.02 |
ENST00000403951.6
|
DGKB
|
diacylglycerol kinase beta |
| chr2_-_157874976 | 0.02 |
ENST00000682025.1
ENST00000683487.1 ENST00000682300.1 ENST00000683441.1 ENST00000684595.1 ENST00000683426.1 ENST00000683820.1 ENST00000263640.7 |
ACVR1
|
activin A receptor type 1 |
| chr14_-_77377046 | 0.02 |
ENST00000216468.8
|
TMED8
|
transmembrane p24 trafficking protein family member 8 |
| chr10_+_91923762 | 0.02 |
ENST00000265990.11
|
BTAF1
|
B-TFIID TATA-box binding protein associated factor 1 |
| chr5_-_95284535 | 0.02 |
ENST00000515393.5
|
MCTP1
|
multiple C2 and transmembrane domain containing 1 |
| chr8_-_138497254 | 0.02 |
ENST00000395297.6
|
FAM135B
|
family with sequence similarity 135 member B |
| chr19_+_33081172 | 0.02 |
ENST00000170564.7
|
GPATCH1
|
G-patch domain containing 1 |
| chr12_+_67648737 | 0.02 |
ENST00000344096.4
ENST00000393555.3 |
DYRK2
|
dual specificity tyrosine phosphorylation regulated kinase 2 |
| chr10_-_119596495 | 0.02 |
ENST00000369093.6
|
TIAL1
|
TIA1 cytotoxic granule associated RNA binding protein like 1 |
| chr3_+_57227714 | 0.02 |
ENST00000288266.8
|
APPL1
|
adaptor protein, phosphotyrosine interacting with PH domain and leucine zipper 1 |
| chr2_-_159798043 | 0.02 |
ENST00000664982.1
ENST00000259053.6 |
ENSG00000287091.1
CD302
|
novel transcript, sense intronic to CD302and LY75-CD302 CD302 molecule |
| chr5_+_112976757 | 0.02 |
ENST00000389063.3
|
DCP2
|
decapping mRNA 2 |
| chr11_-_62727444 | 0.02 |
ENST00000301785.7
|
HNRNPUL2
|
heterogeneous nuclear ribonucleoprotein U like 2 |
| chr11_-_73598183 | 0.02 |
ENST00000064778.8
|
FAM168A
|
family with sequence similarity 168 member A |
| chr1_+_112396200 | 0.02 |
ENST00000271277.11
|
CTTNBP2NL
|
CTTNBP2 N-terminal like |
| chr3_+_152299392 | 0.02 |
ENST00000498502.5
ENST00000545754.5 ENST00000357472.7 ENST00000324196.9 |
MBNL1
|
muscleblind like splicing regulator 1 |
| chr12_-_80937918 | 0.02 |
ENST00000552864.6
|
LIN7A
|
lin-7 homolog A, crumbs cell polarity complex component |
| chr12_+_8697875 | 0.02 |
ENST00000357529.7
|
RIMKLB
|
ribosomal modification protein rimK like family member B |
| chr3_+_108822778 | 0.02 |
ENST00000295756.11
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr1_+_161766309 | 0.02 |
ENST00000679853.1
ENST00000681492.1 ENST00000679886.1 ENST00000367942.4 ENST00000680462.1 ENST00000680633.1 ENST00000681912.1 |
ATF6
|
activating transcription factor 6 |
| chr20_+_2816302 | 0.02 |
ENST00000361033.1
|
TMEM239
|
transmembrane protein 239 |
| chr1_+_26472405 | 0.02 |
ENST00000361427.6
|
HMGN2
|
high mobility group nucleosomal binding domain 2 |
| chr6_+_116571477 | 0.02 |
ENST00000487832.6
ENST00000466444.7 ENST00000518117.5 |
RWDD1
|
RWD domain containing 1 |
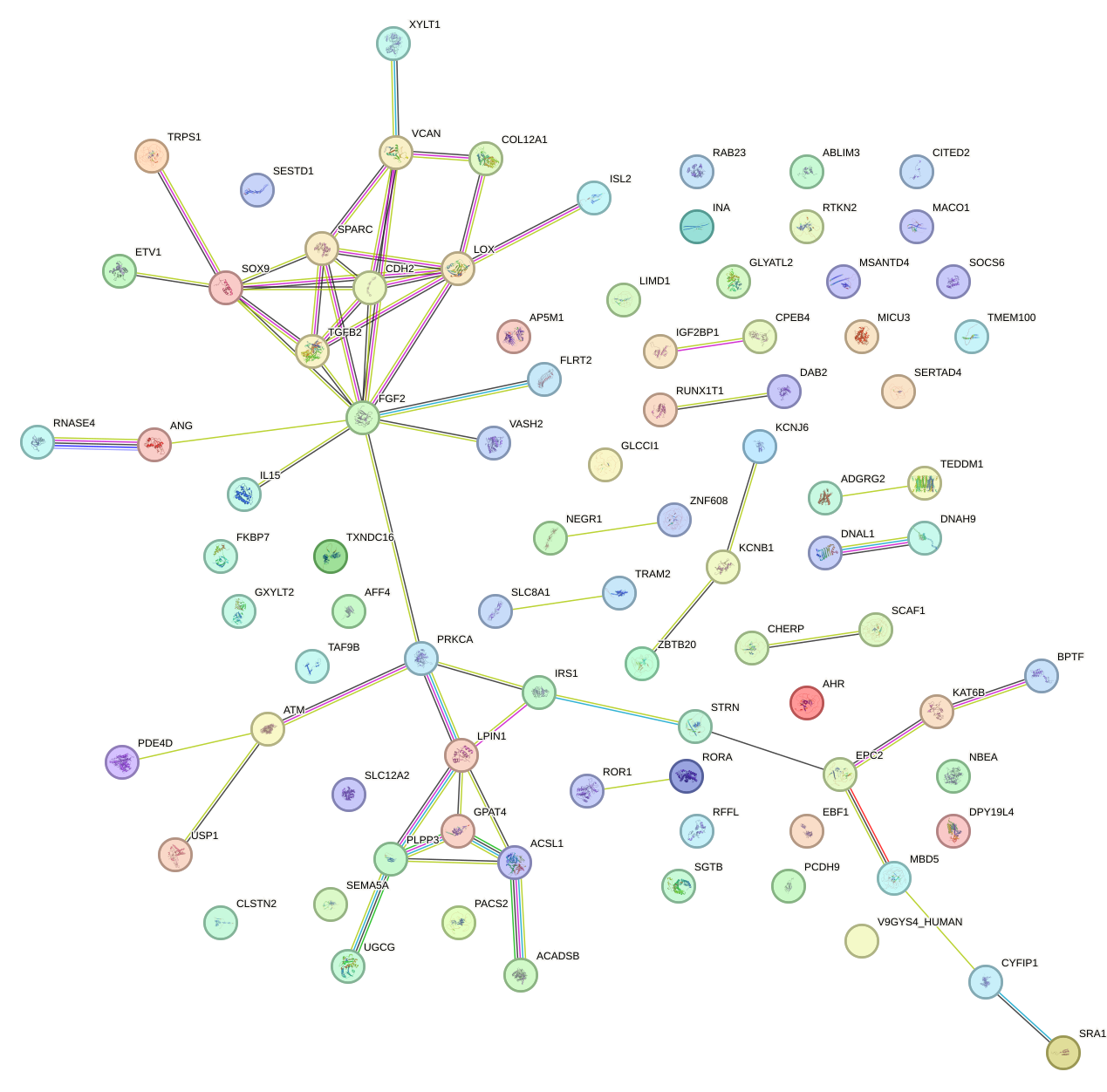
Network of associatons between targets according to the STRING database.
First level regulatory network of UGAAAUG
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.5 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.0 | 0.1 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.0 | 0.4 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:0072434 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.0 | 0.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.1 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.1 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.0 | 0.1 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.1 | GO:0003275 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) negative regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905006) |
| 0.0 | 0.1 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.1 | GO:0006679 | glucosylceramide biosynthetic process(GO:0006679) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.1 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.1 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.0 | 0.1 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.0 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.0 | 0.2 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.0 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.0 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.0 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.0 | 0.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034753 | nuclear aryl hydrocarbon receptor complex(GO:0034753) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0005595 | collagen type XII trimer(GO:0005595) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.1 | 0.4 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.1 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.1 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.0 | 0.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.1 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.0 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.0 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |