Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for UGGCACU
Z-value: 0.84
miRNA associated with seed UGGCACU
| Name | miRBASE accession |
|---|---|
|
hsa-miR-183-5p.2
|
Activity profile of UGGCACU motif
Sorted Z-values of UGGCACU motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_96493803 | 1.01 |
ENST00000518385.5
ENST00000302190.9 |
SDC2
|
syndecan 2 |
| chrX_+_16946650 | 0.72 |
ENST00000357277.8
|
REPS2
|
RALBP1 associated Eps domain containing 2 |
| chr11_+_114059702 | 0.64 |
ENST00000335953.9
ENST00000684612.1 ENST00000682810.1 ENST00000544220.1 |
ZBTB16
|
zinc finger and BTB domain containing 16 |
| chr9_+_4490388 | 0.40 |
ENST00000262352.8
|
SLC1A1
|
solute carrier family 1 member 1 |
| chr7_-_47582076 | 0.40 |
ENST00000311160.14
|
TNS3
|
tensin 3 |
| chr8_+_37796906 | 0.40 |
ENST00000315215.11
|
ADGRA2
|
adhesion G protein-coupled receptor A2 |
| chr16_-_80804581 | 0.39 |
ENST00000570137.7
|
CDYL2
|
chromodomain Y like 2 |
| chr9_+_76459152 | 0.36 |
ENST00000444201.6
ENST00000376730.5 |
GCNT1
|
glucosaminyl (N-acetyl) transferase 1 |
| chr5_-_173328407 | 0.35 |
ENST00000265087.9
|
STC2
|
stanniocalcin 2 |
| chr11_-_83071819 | 0.34 |
ENST00000524635.1
ENST00000526205.5 ENST00000533486.5 ENST00000533276.6 ENST00000527633.6 |
RAB30
|
RAB30, member RAS oncogene family |
| chr12_-_95790755 | 0.30 |
ENST00000343702.9
ENST00000344911.8 |
NTN4
|
netrin 4 |
| chr6_-_57221402 | 0.29 |
ENST00000317483.4
|
RAB23
|
RAB23, member RAS oncogene family |
| chrX_+_9463272 | 0.28 |
ENST00000407597.7
ENST00000380961.5 ENST00000424279.6 |
TBL1X
|
transducin beta like 1 X-linked |
| chr12_-_92145838 | 0.27 |
ENST00000256015.5
|
BTG1
|
BTG anti-proliferation factor 1 |
| chr17_+_48997377 | 0.27 |
ENST00000290341.8
|
IGF2BP1
|
insulin like growth factor 2 mRNA binding protein 1 |
| chr6_+_11537738 | 0.26 |
ENST00000379426.2
|
TMEM170B
|
transmembrane protein 170B |
| chr5_+_68215738 | 0.26 |
ENST00000521381.6
ENST00000521657.5 |
PIK3R1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr8_+_17156463 | 0.25 |
ENST00000262096.13
|
ZDHHC2
|
zinc finger DHHC-type palmitoyltransferase 2 |
| chr11_-_119423162 | 0.22 |
ENST00000284240.10
ENST00000524970.5 |
THY1
|
Thy-1 cell surface antigen |
| chr2_-_226799806 | 0.21 |
ENST00000305123.6
|
IRS1
|
insulin receptor substrate 1 |
| chr12_+_59689337 | 0.21 |
ENST00000261187.8
|
SLC16A7
|
solute carrier family 16 member 7 |
| chr10_+_91220603 | 0.21 |
ENST00000336126.6
|
PCGF5
|
polycomb group ring finger 5 |
| chr2_+_209579399 | 0.21 |
ENST00000360351.8
|
MAP2
|
microtubule associated protein 2 |
| chr7_+_143288215 | 0.20 |
ENST00000619992.4
ENST00000310447.10 |
CASP2
|
caspase 2 |
| chr9_+_88388356 | 0.20 |
ENST00000375859.4
|
SPIN1
|
spindlin 1 |
| chr3_+_138347648 | 0.20 |
ENST00000614350.4
ENST00000289104.8 |
MRAS
|
muscle RAS oncogene homolog |
| chr3_-_9952337 | 0.19 |
ENST00000411976.2
ENST00000412055.6 |
PRRT3
|
proline rich transmembrane protein 3 |
| chr3_-_66500973 | 0.17 |
ENST00000383703.3
ENST00000273261.8 |
LRIG1
|
leucine rich repeats and immunoglobulin like domains 1 |
| chr12_-_48788995 | 0.17 |
ENST00000550422.5
ENST00000357869.8 |
ADCY6
|
adenylate cyclase 6 |
| chr8_+_37762579 | 0.17 |
ENST00000523358.5
ENST00000523187.5 ENST00000328195.8 |
PLPBP
|
pyridoxal phosphate binding protein |
| chrX_-_107716401 | 0.16 |
ENST00000486554.1
ENST00000372390.8 |
TSC22D3
|
TSC22 domain family member 3 |
| chr4_+_76949743 | 0.16 |
ENST00000502584.5
ENST00000264893.11 ENST00000510641.5 |
SEPTIN11
|
septin 11 |
| chr12_-_46825949 | 0.15 |
ENST00000547477.5
ENST00000447411.5 ENST00000266579.9 |
SLC38A4
|
solute carrier family 38 member 4 |
| chr5_+_173888335 | 0.14 |
ENST00000265085.10
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr19_+_32405789 | 0.13 |
ENST00000586987.5
|
DPY19L3
|
dpy-19 like C-mannosyltransferase 3 |
| chr8_+_17497078 | 0.13 |
ENST00000494857.6
ENST00000522656.5 |
SLC7A2
|
solute carrier family 7 member 2 |
| chr5_+_134371561 | 0.13 |
ENST00000265339.7
ENST00000506787.5 ENST00000507277.1 |
UBE2B
|
ubiquitin conjugating enzyme E2 B |
| chr10_+_1049476 | 0.13 |
ENST00000358220.5
|
WDR37
|
WD repeat domain 37 |
| chr5_+_157743703 | 0.13 |
ENST00000286307.6
|
LSM11
|
LSM11, U7 small nuclear RNA associated |
| chr10_+_70404129 | 0.13 |
ENST00000373218.5
|
EIF4EBP2
|
eukaryotic translation initiation factor 4E binding protein 2 |
| chr6_-_52577012 | 0.13 |
ENST00000182527.4
|
TRAM2
|
translocation associated membrane protein 2 |
| chr22_+_30635746 | 0.12 |
ENST00000343605.5
|
SLC35E4
|
solute carrier family 35 member E4 |
| chrX_-_13938618 | 0.12 |
ENST00000454189.6
|
GPM6B
|
glycoprotein M6B |
| chr10_+_61901678 | 0.12 |
ENST00000644638.1
ENST00000681100.1 ENST00000279873.12 |
ARID5B
|
AT-rich interaction domain 5B |
| chr20_-_57710001 | 0.12 |
ENST00000341744.8
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr4_-_113761927 | 0.12 |
ENST00000296402.9
|
CAMK2D
|
calcium/calmodulin dependent protein kinase II delta |
| chr4_-_118352967 | 0.12 |
ENST00000296498.3
|
PRSS12
|
serine protease 12 |
| chr1_-_20486197 | 0.11 |
ENST00000375078.4
|
CAMK2N1
|
calcium/calmodulin dependent protein kinase II inhibitor 1 |
| chr20_+_11890723 | 0.11 |
ENST00000254977.7
|
BTBD3
|
BTB domain containing 3 |
| chr14_-_53152371 | 0.11 |
ENST00000323669.10
|
DDHD1
|
DDHD domain containing 1 |
| chr11_+_45885625 | 0.11 |
ENST00000241014.6
|
MAPK8IP1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr13_+_26557659 | 0.11 |
ENST00000335327.6
ENST00000361042.8 ENST00000671038.1 |
WASF3
|
WASP family member 3 |
| chr7_-_28180735 | 0.11 |
ENST00000283928.10
|
JAZF1
|
JAZF zinc finger 1 |
| chr1_-_38859669 | 0.11 |
ENST00000373001.4
|
RRAGC
|
Ras related GTP binding C |
| chr17_+_68512379 | 0.11 |
ENST00000392711.5
ENST00000585427.5 ENST00000589228.6 ENST00000536854.6 ENST00000588702.5 ENST00000589309.5 |
PRKAR1A
|
protein kinase cAMP-dependent type I regulatory subunit alpha |
| chr5_-_172454487 | 0.10 |
ENST00000311601.6
|
SH3PXD2B
|
SH3 and PX domains 2B |
| chr2_+_45651650 | 0.10 |
ENST00000306156.8
|
PRKCE
|
protein kinase C epsilon |
| chr3_+_152299392 | 0.10 |
ENST00000498502.5
ENST00000545754.5 ENST00000357472.7 ENST00000324196.9 |
MBNL1
|
muscleblind like splicing regulator 1 |
| chr7_+_98617275 | 0.10 |
ENST00000265634.4
|
NPTX2
|
neuronal pentraxin 2 |
| chr6_-_34392627 | 0.10 |
ENST00000607016.2
|
NUDT3
|
nudix hydrolase 3 |
| chr20_-_46406582 | 0.10 |
ENST00000450812.5
ENST00000290246.11 ENST00000396391.5 |
ELMO2
|
engulfment and cell motility 2 |
| chr12_+_27244222 | 0.10 |
ENST00000545470.5
ENST00000389032.8 ENST00000540996.5 |
STK38L
|
serine/threonine kinase 38 like |
| chr2_+_218399838 | 0.10 |
ENST00000273062.7
|
CTDSP1
|
CTD small phosphatase 1 |
| chr12_-_40106026 | 0.10 |
ENST00000280871.9
ENST00000380858.1 |
SLC2A13
|
solute carrier family 2 member 13 |
| chr16_+_8720706 | 0.10 |
ENST00000425191.6
ENST00000569156.5 |
ABAT
|
4-aminobutyrate aminotransferase |
| chr5_-_134632769 | 0.10 |
ENST00000505758.5
ENST00000439578.5 ENST00000502286.1 ENST00000402673.7 |
SAR1B
|
secretion associated Ras related GTPase 1B |
| chr1_+_177170916 | 0.10 |
ENST00000361539.5
|
BRINP2
|
BMP/retinoic acid inducible neural specific 2 |
| chr14_-_74084393 | 0.09 |
ENST00000350259.8
ENST00000553458.6 |
ALDH6A1
|
aldehyde dehydrogenase 6 family member A1 |
| chr3_+_191329342 | 0.09 |
ENST00000392455.9
|
CCDC50
|
coiled-coil domain containing 50 |
| chr17_+_50095331 | 0.09 |
ENST00000503176.6
|
PDK2
|
pyruvate dehydrogenase kinase 2 |
| chr17_-_60526167 | 0.09 |
ENST00000083182.8
|
APPBP2
|
amyloid beta precursor protein binding protein 2 |
| chr3_-_15332526 | 0.09 |
ENST00000383791.8
|
SH3BP5
|
SH3 domain binding protein 5 |
| chr19_+_18097763 | 0.09 |
ENST00000262811.10
|
MAST3
|
microtubule associated serine/threonine kinase 3 |
| chr7_+_17298642 | 0.09 |
ENST00000242057.9
|
AHR
|
aryl hydrocarbon receptor |
| chr3_-_56468346 | 0.09 |
ENST00000288221.11
|
ERC2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr5_+_102755269 | 0.09 |
ENST00000304400.12
ENST00000455264.7 ENST00000684529.1 ENST00000438793.8 ENST00000682882.1 ENST00000682972.1 ENST00000348126.7 ENST00000512073.1 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr2_-_100417608 | 0.09 |
ENST00000264249.8
|
CHST10
|
carbohydrate sulfotransferase 10 |
| chr2_-_134718981 | 0.09 |
ENST00000281924.6
|
TMEM163
|
transmembrane protein 163 |
| chr11_+_120336357 | 0.09 |
ENST00000397843.7
|
ARHGEF12
|
Rho guanine nucleotide exchange factor 12 |
| chr6_-_158818225 | 0.09 |
ENST00000337147.11
|
EZR
|
ezrin |
| chr1_+_111619751 | 0.08 |
ENST00000433097.5
ENST00000369709.3 |
RAP1A
|
RAP1A, member of RAS oncogene family |
| chr9_-_71121596 | 0.08 |
ENST00000377110.9
ENST00000377111.8 ENST00000677713.2 |
TRPM3
|
transient receptor potential cation channel subfamily M member 3 |
| chr9_+_116153783 | 0.08 |
ENST00000328252.4
|
PAPPA
|
pappalysin 1 |
| chr8_-_81112055 | 0.08 |
ENST00000220597.4
|
PAG1
|
phosphoprotein membrane anchor with glycosphingolipid microdomains 1 |
| chrX_+_12975083 | 0.08 |
ENST00000451311.7
ENST00000380636.1 |
TMSB4X
|
thymosin beta 4 X-linked |
| chr3_+_160756225 | 0.08 |
ENST00000498165.6
|
PPM1L
|
protein phosphatase, Mg2+/Mn2+ dependent 1L |
| chr18_+_65751000 | 0.08 |
ENST00000397968.4
|
CDH7
|
cadherin 7 |
| chr1_+_113390495 | 0.08 |
ENST00000307546.14
|
MAGI3
|
membrane associated guanylate kinase, WW and PDZ domain containing 3 |
| chr12_-_109573482 | 0.08 |
ENST00000540016.5
ENST00000545712.7 |
MMAB
|
metabolism of cobalamin associated B |
| chr15_-_50765656 | 0.08 |
ENST00000261854.10
|
SPPL2A
|
signal peptide peptidase like 2A |
| chr3_+_57227714 | 0.08 |
ENST00000288266.8
|
APPL1
|
adaptor protein, phosphotyrosine interacting with PH domain and leucine zipper 1 |
| chr3_-_15859771 | 0.08 |
ENST00000399451.6
|
ANKRD28
|
ankyrin repeat domain 28 |
| chr22_-_28679865 | 0.08 |
ENST00000397906.6
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr9_-_111484353 | 0.07 |
ENST00000338205.9
ENST00000684092.1 |
ECPAS
|
Ecm29 proteasome adaptor and scaffold |
| chr1_-_67833448 | 0.07 |
ENST00000370982.4
|
GNG12
|
G protein subunit gamma 12 |
| chr9_+_2015335 | 0.07 |
ENST00000636559.1
ENST00000349721.8 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr16_-_20900319 | 0.07 |
ENST00000564349.5
ENST00000324344.9 |
ERI2
DCUN1D3
|
ERI1 exoribonuclease family member 2 defective in cullin neddylation 1 domain containing 3 |
| chr8_-_56211257 | 0.07 |
ENST00000316981.8
ENST00000423799.6 ENST00000429357.2 |
PLAG1
|
PLAG1 zinc finger |
| chr3_-_18425295 | 0.07 |
ENST00000338745.11
ENST00000450898.1 |
SATB1
|
SATB homeobox 1 |
| chr6_+_52362088 | 0.07 |
ENST00000635984.1
ENST00000635760.1 ENST00000442253.3 |
EFHC1
PAQR8
|
EF-hand domain containing 1 progestin and adipoQ receptor family member 8 |
| chr11_-_64844620 | 0.07 |
ENST00000342711.6
|
CDC42BPG
|
CDC42 binding protein kinase gamma |
| chr13_-_23375431 | 0.07 |
ENST00000683270.1
ENST00000684163.1 ENST00000402364.1 ENST00000683367.1 |
SACS
|
sacsin molecular chaperone |
| chr16_+_56638659 | 0.07 |
ENST00000290705.12
|
MT1A
|
metallothionein 1A |
| chr1_+_3069160 | 0.07 |
ENST00000511072.5
|
PRDM16
|
PR/SET domain 16 |
| chr13_+_48975879 | 0.07 |
ENST00000492622.6
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr11_+_117178728 | 0.07 |
ENST00000532960.5
ENST00000324225.9 |
SIDT2
|
SID1 transmembrane family member 2 |
| chr17_+_63622406 | 0.07 |
ENST00000579585.5
ENST00000361733.8 ENST00000584573.5 ENST00000361357.7 |
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3 |
| chr5_+_77210667 | 0.06 |
ENST00000264917.10
|
PDE8B
|
phosphodiesterase 8B |
| chrX_+_111096136 | 0.06 |
ENST00000372007.10
|
PAK3
|
p21 (RAC1) activated kinase 3 |
| chr1_+_9292883 | 0.06 |
ENST00000328089.11
|
SPSB1
|
splA/ryanodine receptor domain and SOCS box containing 1 |
| chr5_-_169301098 | 0.06 |
ENST00000519560.6
|
SLIT3
|
slit guidance ligand 3 |
| chr9_-_137028223 | 0.06 |
ENST00000341511.11
|
ABCA2
|
ATP binding cassette subfamily A member 2 |
| chr1_+_117606040 | 0.06 |
ENST00000369448.4
|
TENT5C
|
terminal nucleotidyltransferase 5C |
| chr12_-_54280087 | 0.06 |
ENST00000209875.9
|
CBX5
|
chromobox 5 |
| chr3_+_39809602 | 0.06 |
ENST00000302541.11
ENST00000396217.7 |
MYRIP
|
myosin VIIA and Rab interacting protein |
| chr14_+_55051639 | 0.06 |
ENST00000395468.9
ENST00000622254.1 |
MAPK1IP1L
|
mitogen-activated protein kinase 1 interacting protein 1 like |
| chr15_+_92900189 | 0.06 |
ENST00000626874.2
ENST00000627622.1 ENST00000629346.2 ENST00000628375.2 ENST00000420239.7 ENST00000394196.9 |
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr9_+_32384603 | 0.06 |
ENST00000541043.5
ENST00000379923.5 ENST00000309951.8 |
ACO1
|
aconitase 1 |
| chr18_+_70288991 | 0.06 |
ENST00000397942.4
|
SOCS6
|
suppressor of cytokine signaling 6 |
| chr7_-_27143672 | 0.06 |
ENST00000222726.4
|
HOXA5
|
homeobox A5 |
| chr3_-_39153512 | 0.05 |
ENST00000273153.10
|
CSRNP1
|
cysteine and serine rich nuclear protein 1 |
| chr14_+_54567612 | 0.05 |
ENST00000251091.9
ENST00000392067.7 ENST00000631086.2 |
SAMD4A
|
sterile alpha motif domain containing 4A |
| chr15_-_34754989 | 0.05 |
ENST00000290374.5
|
GJD2
|
gap junction protein delta 2 |
| chr19_+_3094348 | 0.05 |
ENST00000078429.9
|
GNA11
|
G protein subunit alpha 11 |
| chr1_+_7771263 | 0.05 |
ENST00000054666.11
|
VAMP3
|
vesicle associated membrane protein 3 |
| chr9_+_97501622 | 0.05 |
ENST00000259365.9
|
TMOD1
|
tropomodulin 1 |
| chrX_+_73563190 | 0.05 |
ENST00000373504.10
ENST00000373502.9 |
CHIC1
|
cysteine rich hydrophobic domain 1 |
| chr12_-_76559504 | 0.05 |
ENST00000547544.5
ENST00000393249.6 |
OSBPL8
|
oxysterol binding protein like 8 |
| chr2_-_44361754 | 0.05 |
ENST00000409272.5
ENST00000410081.5 ENST00000541738.5 |
PREPL
|
prolyl endopeptidase like |
| chr11_-_6655788 | 0.05 |
ENST00000299441.5
|
DCHS1
|
dachsous cadherin-related 1 |
| chr13_-_30464234 | 0.05 |
ENST00000399489.5
ENST00000339872.8 |
HMGB1
|
high mobility group box 1 |
| chr15_-_75451650 | 0.05 |
ENST00000567289.5
ENST00000394947.8 ENST00000565264.1 |
SIN3A
|
SIN3 transcription regulator family member A |
| chr12_+_110468803 | 0.05 |
ENST00000377673.10
|
FAM216A
|
family with sequence similarity 216 member A |
| chr5_-_172006817 | 0.05 |
ENST00000296933.10
|
FBXW11
|
F-box and WD repeat domain containing 11 |
| chr12_-_89526253 | 0.05 |
ENST00000547474.1
|
POC1B-GALNT4
|
POC1B-GALNT4 readthrough |
| chr17_+_7407838 | 0.05 |
ENST00000302926.7
|
NLGN2
|
neuroligin 2 |
| chr14_+_57268963 | 0.05 |
ENST00000261558.8
|
AP5M1
|
adaptor related protein complex 5 subunit mu 1 |
| chr2_+_20447065 | 0.05 |
ENST00000272233.6
|
RHOB
|
ras homolog family member B |
| chr12_-_118103998 | 0.05 |
ENST00000359236.10
|
VSIG10
|
V-set and immunoglobulin domain containing 10 |
| chrX_-_84188148 | 0.05 |
ENST00000262752.5
|
RPS6KA6
|
ribosomal protein S6 kinase A6 |
| chr7_+_5190196 | 0.05 |
ENST00000401525.7
ENST00000288828.9 ENST00000404704.7 |
WIPI2
|
WD repeat domain, phosphoinositide interacting 2 |
| chr5_+_150508110 | 0.04 |
ENST00000261797.7
|
NDST1
|
N-deacetylase and N-sulfotransferase 1 |
| chr16_+_77788554 | 0.04 |
ENST00000302536.3
|
VAT1L
|
vesicle amine transport 1 like |
| chr3_+_153162196 | 0.04 |
ENST00000323534.5
|
RAP2B
|
RAP2B, member of RAS oncogene family |
| chr1_+_23959797 | 0.04 |
ENST00000374468.1
ENST00000334351.8 |
PNRC2
|
proline rich nuclear receptor coactivator 2 |
| chr1_-_154970735 | 0.04 |
ENST00000368445.9
ENST00000448116.7 ENST00000368449.8 |
SHC1
|
SHC adaptor protein 1 |
| chr9_-_35732122 | 0.04 |
ENST00000314888.10
|
TLN1
|
talin 1 |
| chr20_+_33993646 | 0.04 |
ENST00000375114.7
ENST00000448364.5 |
RALY
|
RALY heterogeneous nuclear ribonucleoprotein |
| chr17_+_32350132 | 0.04 |
ENST00000321233.10
ENST00000394673.6 ENST00000394670.9 ENST00000579634.5 ENST00000580759.5 ENST00000342555.10 ENST00000577908.5 ENST00000394679.9 ENST00000582165.1 |
ZNF207
|
zinc finger protein 207 |
| chr1_-_54887161 | 0.04 |
ENST00000535035.6
ENST00000371269.9 ENST00000436604.2 |
DHCR24
|
24-dehydrocholesterol reductase |
| chr2_+_120252837 | 0.04 |
ENST00000272519.10
ENST00000631312.2 |
RALB
|
RAS like proto-oncogene B |
| chr5_+_65926556 | 0.04 |
ENST00000380943.6
ENST00000416865.6 ENST00000380935.5 ENST00000284037.10 |
ERBIN
|
erbb2 interacting protein |
| chrX_-_19887585 | 0.04 |
ENST00000397821.8
|
SH3KBP1
|
SH3 domain containing kinase binding protein 1 |
| chr20_-_33674359 | 0.04 |
ENST00000606690.5
ENST00000439478.5 ENST00000246190.11 ENST00000375238.8 |
NECAB3
|
N-terminal EF-hand calcium binding protein 3 |
| chr7_+_101085464 | 0.04 |
ENST00000306085.11
ENST00000412507.1 |
TRIM56
|
tripartite motif containing 56 |
| chr5_+_76083360 | 0.04 |
ENST00000502798.7
|
SV2C
|
synaptic vesicle glycoprotein 2C |
| chr14_-_24213446 | 0.04 |
ENST00000530611.1
ENST00000347519.12 ENST00000530996.5 |
ENSG00000254692.1
CHMP4A
|
novel protein charged multivesicular body protein 4A |
| chr2_+_203328378 | 0.04 |
ENST00000430418.5
ENST00000261018.12 ENST00000295851.10 ENST00000424558.5 ENST00000417864.5 ENST00000422511.6 |
ABI2
|
abl interactor 2 |
| chr3_-_72446623 | 0.04 |
ENST00000477973.4
|
RYBP
|
RING1 and YY1 binding protein |
| chr16_+_1678271 | 0.03 |
ENST00000562684.5
ENST00000561516.5 ENST00000248098.8 ENST00000382711.9 ENST00000566742.5 |
JPT2
|
Jupiter microtubule associated homolog 2 |
| chr15_+_63042632 | 0.03 |
ENST00000288398.10
ENST00000358278.7 ENST00000610733.1 ENST00000403994.9 ENST00000357980.9 ENST00000559556.5 ENST00000267996.11 ENST00000559397.6 ENST00000561266.6 ENST00000560970.6 |
TPM1
|
tropomyosin 1 |
| chr13_-_25287457 | 0.03 |
ENST00000381801.6
|
MTMR6
|
myotubularin related protein 6 |
| chr5_+_17217617 | 0.03 |
ENST00000322611.4
|
BASP1
|
brain abundant membrane attached signal protein 1 |
| chr1_-_155562693 | 0.03 |
ENST00000368346.7
ENST00000392403.8 ENST00000679333.1 ENST00000679133.1 |
ASH1L
|
ASH1 like histone lysine methyltransferase |
| chr1_-_153922901 | 0.03 |
ENST00000634401.1
ENST00000368655.5 |
GATAD2B
|
GATA zinc finger domain containing 2B |
| chr19_+_36140059 | 0.03 |
ENST00000246533.8
ENST00000587718.5 ENST00000592483.5 ENST00000590874.5 ENST00000588815.5 |
CAPNS1
|
calpain small subunit 1 |
| chr1_+_35807974 | 0.03 |
ENST00000373210.4
|
AGO4
|
argonaute RISC component 4 |
| chr1_+_201829132 | 0.03 |
ENST00000361565.9
|
IPO9
|
importin 9 |
| chr11_-_77820706 | 0.03 |
ENST00000440064.2
ENST00000528095.5 ENST00000308488.11 |
RSF1
|
remodeling and spacing factor 1 |
| chr20_-_4015389 | 0.03 |
ENST00000336095.10
|
RNF24
|
ring finger protein 24 |
| chr12_+_69470349 | 0.03 |
ENST00000547219.5
ENST00000550316.5 ENST00000548154.5 ENST00000547414.5 ENST00000549921.6 ENST00000550389.5 ENST00000550937.5 ENST00000549092.5 ENST00000550169.5 |
FRS2
|
fibroblast growth factor receptor substrate 2 |
| chr12_-_57237090 | 0.03 |
ENST00000556732.1
|
NDUFA4L2
|
NDUFA4 mitochondrial complex associated like 2 |
| chr1_+_28369705 | 0.03 |
ENST00000373839.8
|
PHACTR4
|
phosphatase and actin regulator 4 |
| chr22_-_38872206 | 0.03 |
ENST00000407418.8
ENST00000216083.6 |
CBX6
|
chromobox 6 |
| chr2_-_160493799 | 0.03 |
ENST00000348849.8
|
RBMS1
|
RNA binding motif single stranded interacting protein 1 |
| chr2_+_46617180 | 0.03 |
ENST00000238892.4
|
CRIPT
|
CXXC repeat containing interactor of PDZ3 domain |
| chr9_+_126860625 | 0.03 |
ENST00000319119.4
|
ZBTB34
|
zinc finger and BTB domain containing 34 |
| chr3_+_115623502 | 0.03 |
ENST00000305124.11
ENST00000393780.3 |
GAP43
|
growth associated protein 43 |
| chr19_+_11355386 | 0.03 |
ENST00000251473.9
ENST00000591329.5 ENST00000586380.5 |
PLPPR2
|
phospholipid phosphatase related 2 |
| chr5_+_132369691 | 0.03 |
ENST00000245407.8
|
SLC22A5
|
solute carrier family 22 member 5 |
| chr1_+_192809031 | 0.03 |
ENST00000235382.7
|
RGS2
|
regulator of G protein signaling 2 |
| chr8_+_42896883 | 0.03 |
ENST00000307602.9
|
HOOK3
|
hook microtubule tethering protein 3 |
| chr8_-_123541197 | 0.03 |
ENST00000517956.5
ENST00000443022.2 |
FBXO32
|
F-box protein 32 |
| chr3_-_147406520 | 0.03 |
ENST00000463250.1
ENST00000383075.8 |
ZIC4
|
Zic family member 4 |
| chr5_+_144205250 | 0.03 |
ENST00000507359.3
|
KCTD16
|
potassium channel tetramerization domain containing 16 |
| chr18_-_26090584 | 0.03 |
ENST00000415083.7
|
SS18
|
SS18 subunit of BAF chromatin remodeling complex |
| chr4_-_77819356 | 0.03 |
ENST00000649644.1
ENST00000504123.6 ENST00000515441.2 |
CNOT6L
|
CCR4-NOT transcription complex subunit 6 like |
| chr12_-_89524734 | 0.03 |
ENST00000529983.3
|
GALNT4
|
polypeptide N-acetylgalactosaminyltransferase 4 |
| chr7_+_107168961 | 0.03 |
ENST00000468410.5
ENST00000478930.5 ENST00000464009.1 ENST00000222574.9 |
HBP1
|
HMG-box transcription factor 1 |
| chr1_-_46668454 | 0.02 |
ENST00000576409.5
|
ATPAF1
|
ATP synthase mitochondrial F1 complex assembly factor 1 |
| chr1_+_99264473 | 0.02 |
ENST00000370185.9
|
PLPPR4
|
phospholipid phosphatase related 4 |
| chrX_-_103688033 | 0.02 |
ENST00000434230.5
ENST00000418819.5 ENST00000360458.5 |
MORF4L2
|
mortality factor 4 like 2 |
| chr7_-_138002017 | 0.02 |
ENST00000452463.5
ENST00000456390.5 ENST00000330387.11 |
CREB3L2
|
cAMP responsive element binding protein 3 like 2 |
| chr14_-_92106607 | 0.02 |
ENST00000340660.10
ENST00000393287.9 ENST00000429774.6 ENST00000545170.5 ENST00000620536.4 ENST00000621269.4 |
ATXN3
|
ataxin 3 |
| chr15_+_50908674 | 0.02 |
ENST00000261842.10
ENST00000560508.1 |
AP4E1
|
adaptor related protein complex 4 subunit epsilon 1 |
| chr12_-_26833143 | 0.02 |
ENST00000381340.8
|
ITPR2
|
inositol 1,4,5-trisphosphate receptor type 2 |
| chr15_+_78872809 | 0.02 |
ENST00000331268.9
|
MORF4L1
|
mortality factor 4 like 1 |
| chr17_+_2796404 | 0.02 |
ENST00000366401.8
ENST00000254695.13 ENST00000542807.1 |
RAP1GAP2
|
RAP1 GTPase activating protein 2 |
| chr9_+_126914760 | 0.02 |
ENST00000424082.6
ENST00000259351.10 ENST00000394022.7 ENST00000394011.7 ENST00000319107.8 |
RALGPS1
|
Ral GEF with PH domain and SH3 binding motif 1 |
| chr5_-_90474765 | 0.02 |
ENST00000316610.7
|
MBLAC2
|
metallo-beta-lactamase domain containing 2 |
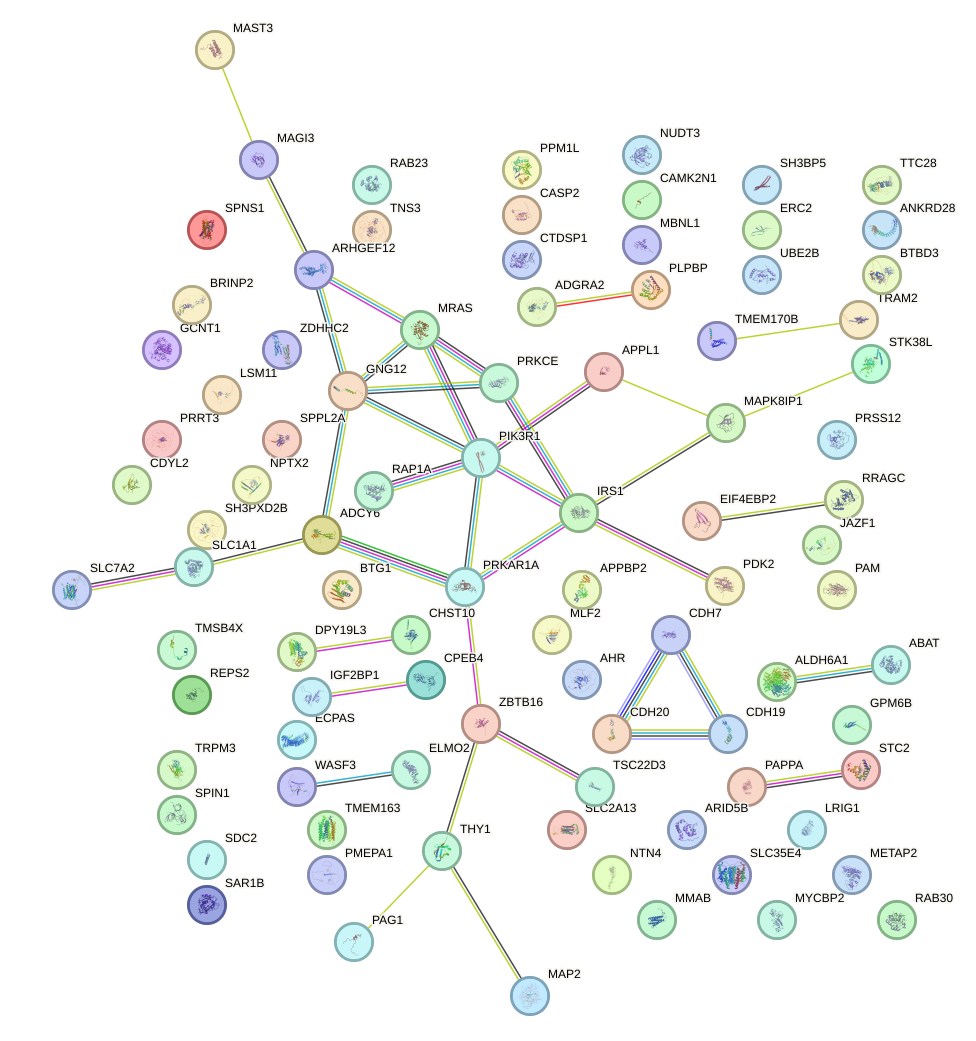
Network of associatons between targets according to the STRING database.
First level regulatory network of UGGCACU
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.6 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.1 | 0.2 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.1 | 0.4 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 0.0 | 0.3 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.2 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:1903400 | L-arginine transmembrane transport(GO:1903400) |
| 0.0 | 0.1 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) positive regulation of meiosis I(GO:0060903) |
| 0.0 | 0.2 | GO:1904117 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.0 | 0.4 | GO:0000101 | sulfur amino acid transport(GO:0000101) |
| 0.0 | 0.4 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.0 | 0.2 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.2 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.0 | 0.1 | GO:1904450 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.0 | 0.1 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.1 | GO:0071543 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.2 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.1 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.3 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.0 | 0.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.1 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.3 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.0 | 0.2 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.1 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) |
| 0.0 | 0.3 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.0 | 0.0 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.0 | GO:0061386 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.1 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.1 | GO:0034753 | nuclear aryl hydrocarbon receptor complex(GO:0034753) |
| 0.0 | 0.1 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.0 | 0.1 | GO:0044393 | microspike(GO:0044393) cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.0 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.0 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.1 | 0.2 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.1 | GO:0005292 | high-affinity basic amino acid transmembrane transporter activity(GO:0005287) high-affinity arginine transmembrane transporter activity(GO:0005289) high-affinity lysine transmembrane transporter activity(GO:0005292) |
| 0.0 | 0.1 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.6 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.4 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.6 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.3 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.1 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0032145 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.0 | 0.1 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.2 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.1 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.0 | 0.1 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.0 | 0.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.1 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.0 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.1 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.4 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |