Project
avrg: Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
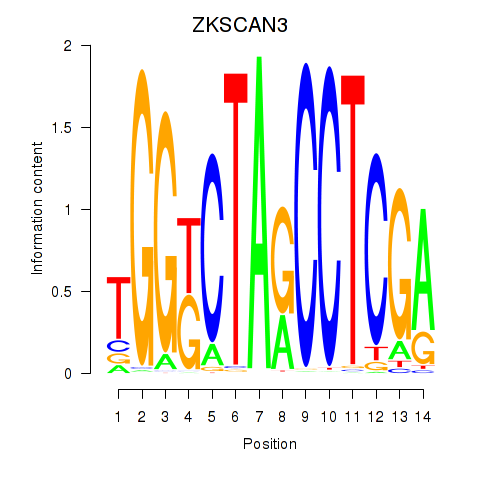
Results for ZKSCAN3
Z-value: 0.73
Transcription factors associated with ZKSCAN3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZKSCAN3
|
ENSG00000189298.14 | zinc finger with KRAB and SCAN domains 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZKSCAN3 | hg38_v1_chr6_+_28349907_28349983 | -0.13 | 7.5e-01 | Click! |
Activity profile of ZKSCAN3 motif
Sorted Z-values of ZKSCAN3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_39296399 | 0.72 |
ENST00000333625.3
|
IFNL1
|
interferon lambda 1 |
| chr21_+_46001300 | 0.47 |
ENST00000612273.2
ENST00000682634.1 |
COL6A1
|
collagen type VI alpha 1 chain |
| chr5_+_176810498 | 0.40 |
ENST00000509580.2
|
UNC5A
|
unc-5 netrin receptor A |
| chr5_+_176810552 | 0.37 |
ENST00000329542.9
|
UNC5A
|
unc-5 netrin receptor A |
| chr2_-_166375969 | 0.30 |
ENST00000454569.6
ENST00000409672.5 |
SCN9A
|
sodium voltage-gated channel alpha subunit 9 |
| chr2_-_166375901 | 0.29 |
ENST00000303354.11
ENST00000645907.1 ENST00000642356.2 ENST00000452182.2 |
SCN9A
|
sodium voltage-gated channel alpha subunit 9 |
| chr2_+_190880809 | 0.24 |
ENST00000320717.8
|
GLS
|
glutaminase |
| chrX_+_86148441 | 0.19 |
ENST00000373125.9
ENST00000373131.5 |
DACH2
|
dachshund family transcription factor 2 |
| chr19_-_39245006 | 0.16 |
ENST00000413851.3
ENST00000613087.4 |
IFNL3
|
interferon lambda 3 |
| chr1_-_19484635 | 0.14 |
ENST00000433834.5
|
CAPZB
|
capping actin protein of muscle Z-line subunit beta |
| chr22_-_49918404 | 0.12 |
ENST00000330817.11
|
ALG12
|
ALG12 alpha-1,6-mannosyltransferase |
| chr14_-_106130061 | 0.11 |
ENST00000390602.3
|
IGHV3-13
|
immunoglobulin heavy variable 3-13 |
| chr5_-_65722094 | 0.11 |
ENST00000381007.9
|
SGTB
|
small glutamine rich tetratricopeptide repeat containing beta |
| chrX_+_151694967 | 0.10 |
ENST00000448726.5
ENST00000538575.5 |
PRRG3
|
proline rich and Gla domain 3 |
| chrX_+_154547606 | 0.09 |
ENST00000594239.6
ENST00000615874.4 ENST00000619941.4 ENST00000617207.4 ENST00000611176.4 |
IKBKG
|
inhibitor of nuclear factor kappa B kinase regulatory subunit gamma |
| chr14_-_80231052 | 0.08 |
ENST00000557010.5
|
DIO2
|
iodothyronine deiodinase 2 |
| chr1_-_19485502 | 0.08 |
ENST00000264203.7
ENST00000375144.6 ENST00000674432.1 ENST00000264202.8 |
CAPZB
|
capping actin protein of muscle Z-line subunit beta |
| chr1_-_19485468 | 0.07 |
ENST00000375142.5
|
CAPZB
|
capping actin protein of muscle Z-line subunit beta |
| chr16_-_695946 | 0.07 |
ENST00000562563.1
|
FBXL16
|
F-box and leucine rich repeat protein 16 |
| chr17_-_18644418 | 0.07 |
ENST00000575220.5
ENST00000405044.6 ENST00000573652.1 |
TBC1D28
|
TBC1 domain family member 28 |
| chr14_-_106675544 | 0.06 |
ENST00000390632.2
|
IGHV3-66
|
immunoglobulin heavy variable 3-66 |
| chr19_+_52369911 | 0.06 |
ENST00000424032.6
ENST00000422689.3 ENST00000600321.5 ENST00000344085.9 ENST00000597976.5 |
ZNF880
|
zinc finger protein 880 |
| chr3_+_50279080 | 0.05 |
ENST00000316436.4
|
LSMEM2
|
leucine rich single-pass membrane protein 2 |
| chr17_+_38752731 | 0.05 |
ENST00000619426.5
ENST00000610434.4 |
PSMB3
|
proteasome 20S subunit beta 3 |
| chr5_+_140700437 | 0.05 |
ENST00000274712.8
|
ZMAT2
|
zinc finger matrin-type 2 |
| chr14_-_106593319 | 0.04 |
ENST00000390627.3
|
IGHV3-53
|
immunoglobulin heavy variable 3-53 |
| chr15_+_74318553 | 0.04 |
ENST00000558821.5
ENST00000268082.4 |
CCDC33
|
coiled-coil domain containing 33 |
| chr1_+_93448108 | 0.04 |
ENST00000271234.13
ENST00000260506.12 |
FNBP1L
|
formin binding protein 1 like |
| chr16_-_28495519 | 0.04 |
ENST00000569430.7
|
CLN3
|
CLN3 lysosomal/endosomal transmembrane protein, battenin |
| chr5_-_115262851 | 0.04 |
ENST00000379615.3
ENST00000419445.6 |
PGGT1B
|
protein geranylgeranyltransferase type I subunit beta |
| chr12_+_71754834 | 0.04 |
ENST00000261263.5
|
RAB21
|
RAB21, member RAS oncogene family |
| chr1_+_93448155 | 0.03 |
ENST00000370253.6
|
FNBP1L
|
formin binding protein 1 like |
| chr1_+_226062704 | 0.03 |
ENST00000366814.3
ENST00000366815.10 ENST00000655399.1 ENST00000667897.1 |
H3-3A
|
H3.3 histone A |
| chr11_-_64246190 | 0.02 |
ENST00000392210.6
|
PPP1R14B
|
protein phosphatase 1 regulatory inhibitor subunit 14B |
| chr14_+_64113084 | 0.02 |
ENST00000673797.1
|
SYNE2
|
spectrin repeat containing nuclear envelope protein 2 |
| chr7_+_1044542 | 0.02 |
ENST00000444847.2
|
GPR146
|
G protein-coupled receptor 146 |
| chr22_+_49918626 | 0.02 |
ENST00000328268.9
ENST00000404488.7 |
CRELD2
|
cysteine rich with EGF like domains 2 |
| chr11_-_119346695 | 0.02 |
ENST00000619721.6
|
MFRP
|
membrane frizzled-related protein |
| chr5_+_65722190 | 0.01 |
ENST00000380985.10
ENST00000502464.5 |
NLN
|
neurolysin |
| chr11_-_61295289 | 0.01 |
ENST00000335613.10
|
VWCE
|
von Willebrand factor C and EGF domains |
| chr6_-_31540536 | 0.01 |
ENST00000376177.6
|
DDX39B
|
DExD-box helicase 39B |
| chr7_+_155298561 | 0.01 |
ENST00000476756.1
|
INSIG1
|
insulin induced gene 1 |
| chr14_+_64113054 | 0.01 |
ENST00000673869.1
|
SYNE2
|
spectrin repeat containing nuclear envelope protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZKSCAN3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.1 | 0.8 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.3 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 0.6 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.5 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.0 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.0 | 0.5 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.8 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.0 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.6 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.0 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.0 | 0.2 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |