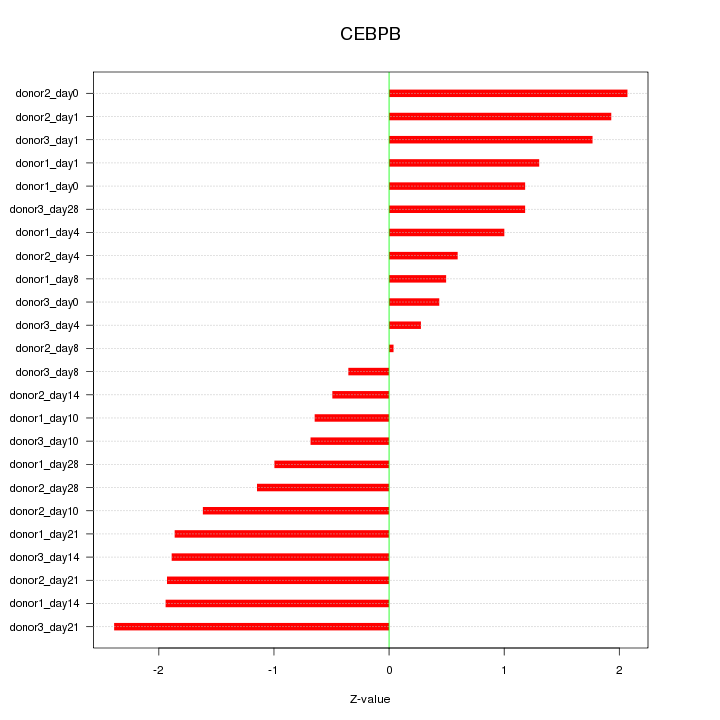
Motif ID: CEBPB
Z-value: 1.353

Transcription factors associated with CEBPB:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| CEBPB | ENSG00000172216.4 | CEBPB |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CEBPB | hg19_v2_chr20_+_48807351_48807384 | -0.22 | 3.0e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.9 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) |
| 1.4 | 9.5 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 1.3 | 7.6 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 1.2 | 3.7 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 1.1 | 4.3 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.9 | 2.6 | GO:0070512 | regulation of histone H4-K20 methylation(GO:0070510) positive regulation of histone H4-K20 methylation(GO:0070512) |
| 0.7 | 15.3 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.7 | 2.1 | GO:0033242 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.7 | 3.4 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.6 | 5.6 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.6 | 1.8 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.6 | 2.5 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.6 | 1.8 | GO:0006423 | cysteinyl-tRNA aminoacylation(GO:0006423) |
| 0.5 | 1.9 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.4 | 2.7 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.4 | 2.5 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.4 | 1.7 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.4 | 1.6 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.4 | 1.5 | GO:1902613 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.4 | 1.1 | GO:0045994 | positive regulation of translational initiation by iron(GO:0045994) |
| 0.3 | 1.7 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.3 | 2.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.3 | 1.6 | GO:0060356 | leucine import(GO:0060356) |
| 0.3 | 2.2 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.3 | 1.2 | GO:0072255 | metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 0.3 | 1.6 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.3 | 0.8 | GO:2000547 | regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.2 | 1.5 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.2 | 1.5 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.2 | 2.1 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.2 | 0.9 | GO:0036229 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.2 | 0.7 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.2 | 0.6 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.2 | 1.0 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.2 | 0.6 | GO:0060178 | regulation of exocyst assembly(GO:0001928) regulation of exocyst localization(GO:0060178) |
| 0.2 | 0.8 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 0.2 | 0.9 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.2 | 0.9 | GO:0019556 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.2 | 1.6 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.2 | 1.1 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.2 | 0.5 | GO:0006424 | glutamyl-tRNA aminoacylation(GO:0006424) prolyl-tRNA aminoacylation(GO:0006433) |
| 0.2 | 0.8 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) regulation of determination of dorsal identity(GO:2000015) |
| 0.2 | 3.8 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.2 | 0.8 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.2 | 4.7 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 0.6 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.1 | 1.8 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.1 | 4.5 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.1 | 0.6 | GO:1904640 | response to methionine(GO:1904640) |
| 0.1 | 0.3 | GO:0098759 | response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) |
| 0.1 | 0.5 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.4 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.1 | 0.3 | GO:0042495 | detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.1 | 1.1 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) negative regulation of smooth muscle cell apoptotic process(GO:0034392) negative regulation of fibrinolysis(GO:0051918) |
| 0.1 | 0.7 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 | 0.6 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.1 | 0.5 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.1 | 0.2 | GO:1990258 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.1 | 0.5 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.1 | 0.3 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.1 | 1.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.6 | GO:0036093 | male germ cell proliferation(GO:0002176) germ cell proliferation(GO:0036093) |
| 0.1 | 0.6 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.1 | 0.2 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.1 | 0.3 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.1 | 1.2 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.1 | 0.3 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.1 | 0.4 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.1 | 0.3 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 0.8 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.1 | 0.5 | GO:1903912 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.0 | 0.4 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.1 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.0 | 0.9 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.5 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.2 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.2 | GO:0052199 | response to yeast(GO:0001878) negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.0 | 0.2 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.4 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.3 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.0 | 2.4 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.2 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 4.0 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.4 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 1.6 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.6 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 2.5 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.5 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.0 | 0.4 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.3 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.2 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.6 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 1.0 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.7 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.5 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.0 | 0.6 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.0 | 1.8 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.1 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 | 0.1 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 9.5 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.8 | 2.5 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.6 | 2.6 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 0.3 | 4.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 1.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.2 | 1.0 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.2 | 1.9 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 0.3 | GO:0034665 | integrin alpha1-beta1 complex(GO:0034665) |
| 0.1 | 1.5 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 0.8 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 0.3 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.1 | 2.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.5 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 2.1 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.1 | 2.1 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 6.3 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.5 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 1.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.6 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.3 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.3 | GO:1904813 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 1.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.2 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.0 | 0.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 4.6 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 3.6 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.4 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.8 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 3.0 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.8 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.5 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 2.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 7.2 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.4 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 1.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 3.0 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.4 | GO:0016592 | mediator complex(GO:0016592) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.6 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 1.6 | 4.9 | GO:0050421 | cystathionine beta-synthase activity(GO:0004122) oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) nitrite reductase (NO-forming) activity(GO:0050421) carbon monoxide binding(GO:0070025) nitrite reductase activity(GO:0098809) |
| 1.6 | 8.0 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 1.4 | 9.5 | GO:0008732 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 1.3 | 7.6 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 1.1 | 4.3 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.8 | 15.3 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.7 | 2.1 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.7 | 3.4 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.6 | 2.5 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.6 | 1.8 | GO:0004817 | cysteine-tRNA ligase activity(GO:0004817) |
| 0.6 | 1.7 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.5 | 2.5 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.4 | 1.2 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.4 | 2.2 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.4 | 1.1 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.3 | 2.7 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.3 | 2.0 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.3 | 0.5 | GO:0070905 | serine binding(GO:0070905) |
| 0.3 | 2.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.3 | 1.9 | GO:0070728 | leucine binding(GO:0070728) |
| 0.3 | 0.8 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.3 | 1.6 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.2 | 2.8 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.2 | 4.7 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.2 | 0.6 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.2 | 0.8 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.2 | 2.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.2 | 1.5 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.2 | 0.5 | GO:0004827 | glutamate-tRNA ligase activity(GO:0004818) proline-tRNA ligase activity(GO:0004827) |
| 0.2 | 0.8 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 1.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.6 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.1 | 0.7 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.1 | 0.9 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.1 | 0.3 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.1 | 1.7 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 1.6 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 1.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.3 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.1 | 1.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.2 | GO:1990259 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.1 | 0.4 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.1 | 0.5 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.3 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.1 | 1.5 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 3.7 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.1 | 0.3 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 1.3 | GO:0015464 | acetylcholine receptor activity(GO:0015464) |
| 0.0 | 0.8 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.5 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 3.1 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 1.8 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.2 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 1.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 1.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.2 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.4 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.5 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.2 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 2.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 1.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.7 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.5 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 2.0 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.4 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.5 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.7 | PID_INTEGRIN5_PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 5.6 | PID_IL1_PATHWAY | IL1-mediated signaling events |
| 0.1 | 3.4 | PID_RXR_VDR_PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 4.9 | PID_FRA_PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 6.9 | PID_MYC_ACTIV_PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 2.6 | PID_BARD1_PATHWAY | BARD1 signaling events |
| 0.0 | 4.7 | PID_BCR_5PATHWAY | BCR signaling pathway |
| 0.0 | 2.0 | PID_P38_ALPHA_BETA_PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.2 | PID_S1P_S1P1_PATHWAY | S1P1 pathway |
| 0.0 | 1.8 | PID_IGF1_PATHWAY | IGF1 pathway |
| 0.0 | 1.1 | PID_REELIN_PATHWAY | Reelin signaling pathway |
| 0.0 | 0.3 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.6 | PID_NFAT_TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.3 | PID_NFKAPPAB_CANONICAL_PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.6 | ST_INTERLEUKIN_4_PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.3 | PID_LYMPH_ANGIOGENESIS_PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 1.0 | PID_TNF_PATHWAY | TNF receptor signaling pathway |
| 0.0 | 1.5 | PID_ECADHERIN_STABILIZATION_PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.0 | PID_CDC42_REG_PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.3 | PID_EPHRINB_REV_PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.5 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.8 | PID_P38_ALPHA_BETA_DOWNSTREAM_PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.4 | PID_CONE_PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.2 | PID_IL27_PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.8 | PID_P53_REGULATION_PATHWAY | p53 pathway |
| 0.0 | 0.3 | PID_SYNDECAN_2_PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.3 | PID_TOLL_ENDOGENOUS_PATHWAY | Endogenous TLR signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 15.3 | REACTOME_NEGATIVE_REGULATION_OF_THE_PI3K_AKT_NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.3 | 12.9 | REACTOME_BASIGIN_INTERACTIONS | Genes involved in Basigin interactions |
| 0.3 | 7.6 | REACTOME_BRANCHED_CHAIN_AMINO_ACID_CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 8.0 | REACTOME_CYTOSOLIC_TRNA_AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 2.8 | REACTOME_APOPTOTIC_CLEAVAGE_OF_CELL_ADHESION_PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 3.8 | REACTOME_AMINO_ACID_SYNTHESIS_AND_INTERCONVERSION_TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.8 | REACTOME_SIGNAL_ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 5.0 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 3.5 | REACTOME_GRB2_SOS_PROVIDES_LINKAGE_TO_MAPK_SIGNALING_FOR_INTERGRINS_ | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 2.6 | REACTOME_AMINO_ACID_TRANSPORT_ACROSS_THE_PLASMA_MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 2.6 | REACTOME_HOMOLOGOUS_RECOMBINATION_REPAIR_OF_REPLICATION_INDEPENDENT_DOUBLE_STRAND_BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 5.6 | REACTOME_IL1_SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 1.9 | REACTOME_NA_CL_DEPENDENT_NEUROTRANSMITTER_TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 0.8 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_ATF6_ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.8 | REACTOME_RECYCLING_OF_BILE_ACIDS_AND_SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 1.7 | REACTOME_ASSOCIATION_OF_TRIC_CCT_WITH_TARGET_PROTEINS_DURING_BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.4 | REACTOME_NOREPINEPHRINE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME_CLASS_C_3_METABOTROPIC_GLUTAMATE_PHEROMONE_RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.3 | REACTOME_REGULATION_OF_INSULIN_SECRETION_BY_ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.6 | REACTOME_P38MAPK_EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.3 | REACTOME_CDC6_ASSOCIATION_WITH_THE_ORC_ORIGIN_COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 4.4 | REACTOME_G_ALPHA_I_SIGNALLING_EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 1.1 | REACTOME_FORMATION_OF_THE_TERNARY_COMPLEX_AND_SUBSEQUENTLY_THE_43S_COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.3 | REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.6 | REACTOME_DOWNREGULATION_OF_TGF_BETA_RECEPTOR_SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 1.2 | REACTOME_DOWNSTREAM_SIGNAL_TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 1.9 | REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.3 | REACTOME_EXTRINSIC_PATHWAY_FOR_APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.9 | REACTOME_INTERFERON_ALPHA_BETA_SIGNALING | Genes involved in Interferon alpha/beta signaling |