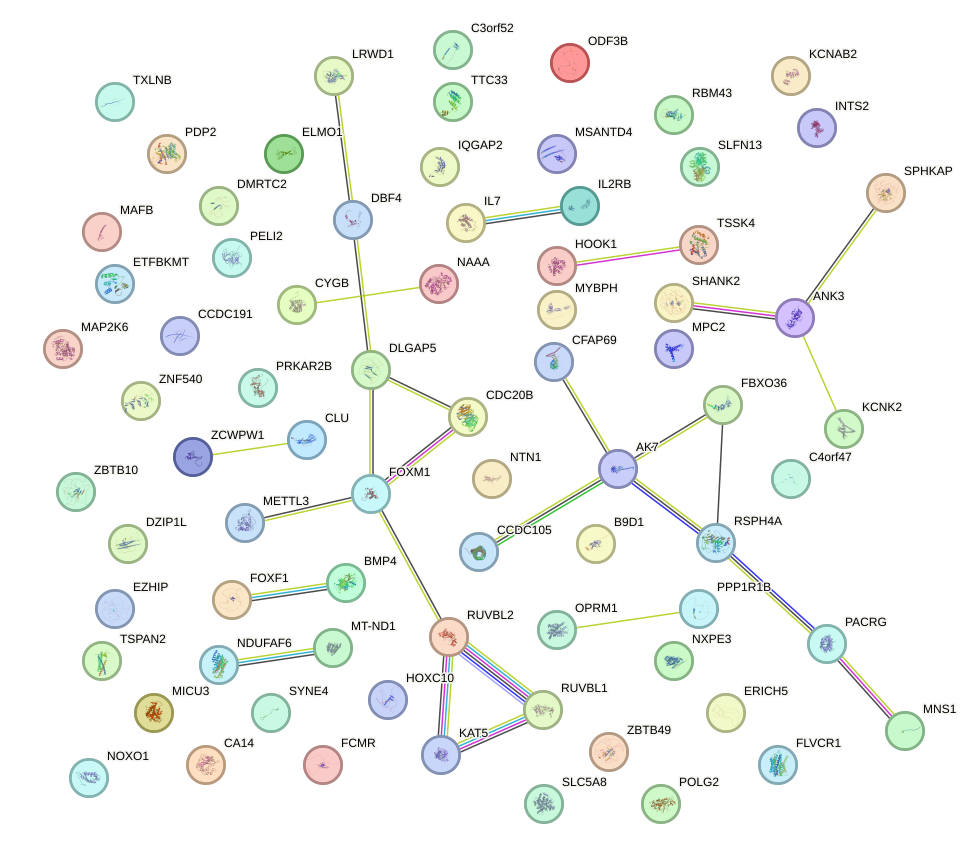
| 0.3 |
1.3 |
GO:2000768 |
glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.2 |
1.0 |
GO:1901842 |
negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 |
0.6 |
GO:1902361 |
mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.1 |
0.7 |
GO:0007079 |
mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.1 |
0.5 |
GO:0090202 |
transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.1 |
0.3 |
GO:0072709 |
cellular response to sorbitol(GO:0072709) |
| 0.1 |
0.3 |
GO:0007439 |
ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) right lung development(GO:0060458) |
| 0.1 |
0.3 |
GO:1905072 |
apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.1 |
0.3 |
GO:1900039 |
positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.1 |
0.3 |
GO:0035283 |
rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 |
0.9 |
GO:0051388 |
positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 |
0.2 |
GO:1900111 |
positive regulation of histone H3-K9 dimethylation(GO:1900111) |
| 0.1 |
0.3 |
GO:0051892 |
negative regulation of cardioblast differentiation(GO:0051892) regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.1 |
0.5 |
GO:2000490 |
negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.1 |
0.5 |
GO:0038110 |
interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.1 |
0.3 |
GO:0086021 |
SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.1 |
0.4 |
GO:0044208 |
'de novo' AMP biosynthetic process(GO:0044208) |
| 0.1 |
0.4 |
GO:0006382 |
adenosine to inosine editing(GO:0006382) |
| 0.1 |
0.6 |
GO:0000492 |
box C/D snoRNP assembly(GO:0000492) |
| 0.1 |
0.3 |
GO:2001170 |
negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.1 |
0.4 |
GO:0014005 |
microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.1 |
0.1 |
GO:0032764 |
negative regulation of mast cell cytokine production(GO:0032764) |
| 0.1 |
0.3 |
GO:0031860 |
telomeric 3' overhang formation(GO:0031860) |
| 0.1 |
0.2 |
GO:0060086 |
circadian temperature homeostasis(GO:0060086) positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.1 |
0.2 |
GO:0061358 |
negative regulation of Wnt protein secretion(GO:0061358) |
| 0.1 |
1.1 |
GO:0070986 |
left/right axis specification(GO:0070986) |
| 0.1 |
0.2 |
GO:0003249 |
left ventricular cardiac muscle tissue morphogenesis(GO:0003220) cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) |
| 0.1 |
0.2 |
GO:0035669 |
TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.1 |
0.4 |
GO:0071169 |
establishment of protein localization to chromatin(GO:0071169) |
| 0.1 |
0.7 |
GO:0070070 |
proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 |
0.2 |
GO:0072658 |
maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 |
0.1 |
GO:0036353 |
histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 |
3.0 |
GO:0035082 |
axoneme assembly(GO:0035082) |
| 0.0 |
0.2 |
GO:0071484 |
response to high light intensity(GO:0009644) cellular response to light intensity(GO:0071484) |
| 0.0 |
0.1 |
GO:0042450 |
arginine biosynthetic process via ornithine(GO:0042450) |
| 0.0 |
0.1 |
GO:0032240 |
mRNA export from nucleus in response to heat stress(GO:0031990) negative regulation of nucleobase-containing compound transport(GO:0032240) positive regulation of chromatin assembly or disassembly(GO:0045799) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.0 |
0.3 |
GO:0097338 |
response to clozapine(GO:0097338) |
| 0.0 |
0.1 |
GO:0071557 |
regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) histone H3-K27 demethylation(GO:0071557) |
| 0.0 |
0.1 |
GO:0052151 |
positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.0 |
0.2 |
GO:0097089 |
methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.0 |
0.2 |
GO:1902998 |
macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.0 |
0.3 |
GO:0016191 |
synaptic vesicle uncoating(GO:0016191) |
| 0.0 |
0.1 |
GO:0006424 |
glutamyl-tRNA aminoacylation(GO:0006424) |
| 0.0 |
0.1 |
GO:0046081 |
dUTP metabolic process(GO:0046080) dUTP catabolic process(GO:0046081) |
| 0.0 |
0.2 |
GO:0008063 |
Toll signaling pathway(GO:0008063) |
| 0.0 |
0.2 |
GO:0038018 |
Wnt receptor catabolic process(GO:0038018) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 |
0.4 |
GO:0015886 |
heme transport(GO:0015886) |
| 0.0 |
0.2 |
GO:0033564 |
anterior/posterior axon guidance(GO:0033564) |
| 0.0 |
0.1 |
GO:0097156 |
fasciculation of motor neuron axon(GO:0097156) |
| 0.0 |
0.1 |
GO:0006154 |
adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 0.0 |
0.1 |
GO:0097195 |
pilomotor reflex(GO:0097195) |
| 0.0 |
0.2 |
GO:0007621 |
negative regulation of female receptivity(GO:0007621) |
| 0.0 |
0.4 |
GO:0035269 |
protein O-linked mannosylation(GO:0035269) |
| 0.0 |
0.1 |
GO:0045829 |
negative regulation of isotype switching(GO:0045829) |
| 0.0 |
0.2 |
GO:0017185 |
peptidyl-lysine hydroxylation(GO:0017185) |
| 0.0 |
0.1 |
GO:1902683 |
regulation of receptor localization to synapse(GO:1902683) |
| 0.0 |
0.1 |
GO:0019243 |
methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 |
0.6 |
GO:0034587 |
piRNA metabolic process(GO:0034587) |
| 0.0 |
0.3 |
GO:0021520 |
spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 |
1.0 |
GO:0010738 |
regulation of protein kinase A signaling(GO:0010738) |
| 0.0 |
0.7 |
GO:0032703 |
negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 |
0.6 |
GO:0070493 |
thrombin receptor signaling pathway(GO:0070493) |
| 0.0 |
0.2 |
GO:0032306 |
regulation of prostaglandin secretion(GO:0032306) positive regulation of prostaglandin secretion(GO:0032308) |
| 0.0 |
0.2 |
GO:0045198 |
establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 |
0.1 |
GO:0007113 |
endomitotic cell cycle(GO:0007113) thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 |
0.2 |
GO:0015840 |
urea transport(GO:0015840) |
| 0.0 |
1.7 |
GO:0042733 |
embryonic digit morphogenesis(GO:0042733) |
| 0.0 |
1.4 |
GO:0015949 |
nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 |
0.1 |
GO:0072053 |
renal inner medulla development(GO:0072053) renal outer medulla development(GO:0072054) inner medullary collecting duct development(GO:0072061) |
| 0.0 |
0.1 |
GO:1902202 |
regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 |
0.3 |
GO:0030422 |
production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 |
0.1 |
GO:0021784 |
postganglionic parasympathetic fiber development(GO:0021784) regulation of ureteric bud formation(GO:0072106) positive regulation of ureteric bud formation(GO:0072107) |
| 0.0 |
1.3 |
GO:0010501 |
RNA secondary structure unwinding(GO:0010501) |
| 0.0 |
0.4 |
GO:0060263 |
regulation of respiratory burst(GO:0060263) |
| 0.0 |
0.1 |
GO:0071317 |
cellular response to morphine(GO:0071315) cellular response to isoquinoline alkaloid(GO:0071317) |
| 0.0 |
0.4 |
GO:0050812 |
acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 |
0.2 |
GO:0006390 |
transcription from mitochondrial promoter(GO:0006390) |
| 0.0 |
0.4 |
GO:0071688 |
striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 |
0.1 |
GO:0090116 |
C-5 methylation of cytosine(GO:0090116) |
| 0.0 |
0.2 |
GO:0009048 |
dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 |
0.1 |
GO:0038188 |
cholecystokinin signaling pathway(GO:0038188) |
| 0.0 |
0.1 |
GO:0015798 |
myo-inositol transport(GO:0015798) |
| 0.0 |
0.2 |
GO:0052805 |
histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 |
0.1 |
GO:0035964 |
COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 |
0.1 |
GO:0019368 |
fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 |
0.1 |
GO:0007060 |
male meiosis chromosome segregation(GO:0007060) |
| 0.0 |
0.1 |
GO:0022027 |
interkinetic nuclear migration(GO:0022027) |
| 0.0 |
0.2 |
GO:0042996 |
regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 |
0.1 |
GO:1903608 |
protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 |
0.1 |
GO:0035711 |
plasmacytoid dendritic cell activation(GO:0002270) T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) regulation of restriction endodeoxyribonuclease activity(GO:0032072) T-helper 1 cell activation(GO:0035711) |
| 0.0 |
0.1 |
GO:0000973 |
posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 |
0.1 |
GO:0070966 |
nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 |
0.1 |
GO:0016198 |
axon choice point recognition(GO:0016198) |
| 0.0 |
0.1 |
GO:0019626 |
short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 |
0.2 |
GO:0034472 |
snRNA 3'-end processing(GO:0034472) |
| 0.0 |
0.1 |
GO:0030644 |
cellular chloride ion homeostasis(GO:0030644) |
| 0.0 |
0.1 |
GO:0071392 |
cellular response to estradiol stimulus(GO:0071392) |
| 0.0 |
0.5 |
GO:0032878 |
regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 |
0.1 |
GO:0051182 |
coenzyme transport(GO:0051182) |
| 0.0 |
0.1 |
GO:0000480 |
endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 |
0.1 |
GO:0007498 |
mesoderm development(GO:0007498) |
| 0.0 |
0.1 |
GO:1901621 |
negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 |
0.3 |
GO:0003334 |
keratinocyte development(GO:0003334) |
| 0.0 |
0.1 |
GO:0061084 |
regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.0 |
0.0 |
GO:0060214 |
endocardium formation(GO:0060214) |
| 0.0 |
0.4 |
GO:0060292 |
long term synaptic depression(GO:0060292) |
| 0.0 |
0.1 |
GO:0051673 |
membrane disruption in other organism(GO:0051673) |
| 0.0 |
0.2 |
GO:0006048 |
UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 |
0.1 |
GO:0060075 |
regulation of resting membrane potential(GO:0060075) |
| 0.0 |
0.1 |
GO:0072498 |
embryonic skeletal joint development(GO:0072498) |
| 0.0 |
0.1 |
GO:0006620 |
posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 |
0.2 |
GO:0032042 |
mitochondrial DNA metabolic process(GO:0032042) |
| 0.0 |
0.2 |
GO:0007342 |
fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 |
0.0 |
GO:1900194 |
negative regulation of oocyte maturation(GO:1900194) |
| 0.0 |
0.1 |
GO:0070934 |
CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 |
0.3 |
GO:0035988 |
chondrocyte proliferation(GO:0035988) |
| 0.0 |
0.2 |
GO:0034356 |
NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 |
0.1 |
GO:0031053 |
primary miRNA processing(GO:0031053) |
| 0.0 |
0.1 |
GO:0006044 |
N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 |
0.2 |
GO:0060044 |
negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.0 |
0.3 |
GO:0046475 |
glycerophospholipid catabolic process(GO:0046475) |