Motif ID: HOXD4
Z-value: 0.377
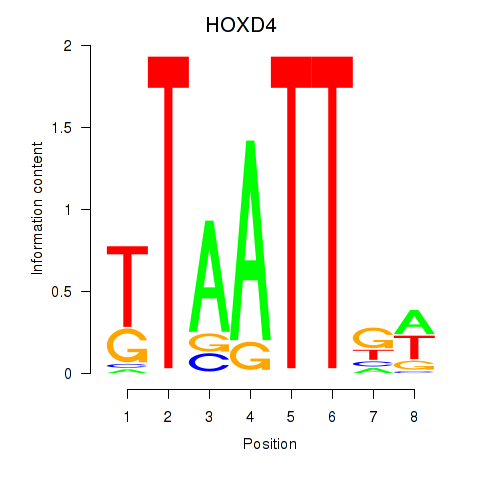
Transcription factors associated with HOXD4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| HOXD4 | ENSG00000170166.5 | HOXD4 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXD4 | hg19_v2_chr2_+_177015950_177015950 | -0.37 | 7.2e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 0.1 | 0.5 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.1 | 0.3 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.3 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.1 | 0.3 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.1 | 0.1 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.1 | 0.3 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.2 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.1 | 0.7 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.9 | GO:0036158 | lateral ventricle development(GO:0021670) outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.2 | GO:0051958 | methotrexate transport(GO:0051958) |
| 0.0 | 0.2 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.4 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 0.2 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.2 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.3 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.1 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.2 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.0 | GO:0099543 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.1 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.0 | 0.1 | GO:0090425 | hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.0 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.4 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.0 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.1 | 0.9 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.2 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.3 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.3 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.1 | 0.3 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 0.3 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.1 | 0.2 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.5 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.9 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.2 | GO:0015350 | methotrexate transporter activity(GO:0015350) |
| 0.0 | 0.4 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.2 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.0 | 0.2 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.1 | GO:0097363 | protein N-acetylglucosaminyltransferase activity(GO:0016262) protein O-GlcNAc transferase activity(GO:0097363) |
| 0.0 | 0.1 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 0.7 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.2 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID_SYNDECAN_3_PATHWAY | Syndecan-3-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME_ANDROGEN_BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.4 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |





