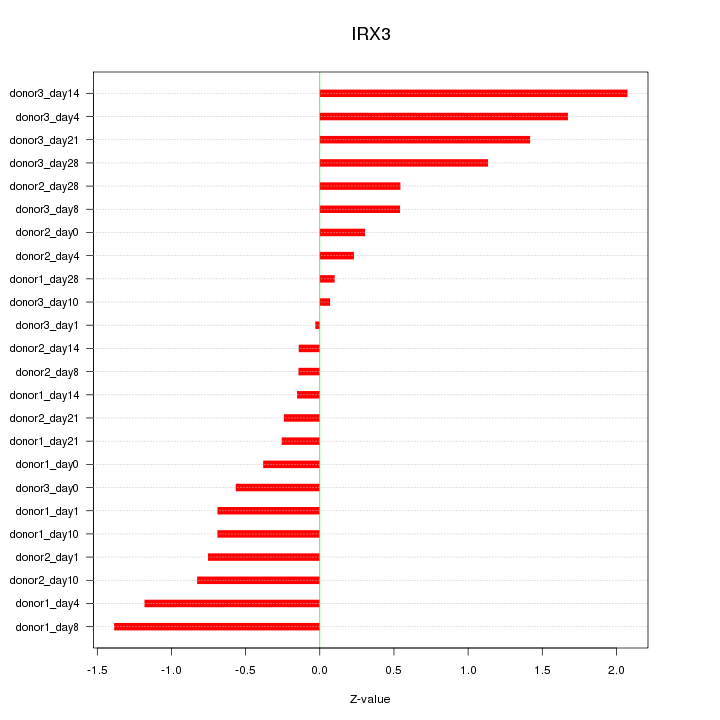
Motif ID: IRX3
Z-value: 0.850

Transcription factors associated with IRX3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| IRX3 | ENSG00000177508.11 | IRX3 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRX3 | hg19_v2_chr16_-_54320675_54320715 | 0.04 | 8.4e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.2 | 3.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.2 | 0.6 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.2 | 0.8 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.1 | 0.4 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.1 | 0.9 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 0.3 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.1 | 0.3 | GO:0002818 | intracellular defense response(GO:0002818) |
| 0.1 | 2.3 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.1 | 0.2 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.1 | 0.5 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.1 | 1.0 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 1.9 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 0.6 | GO:0002784 | regulation of antimicrobial peptide production(GO:0002784) regulation of antibacterial peptide production(GO:0002786) negative regulation of defense response to bacterium(GO:1900425) |
| 0.1 | 0.7 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.1 | 0.6 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 1.1 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.1 | 1.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 0.6 | GO:2000334 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 0.2 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.1 | 0.2 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.1 | 0.4 | GO:1990523 | response to vitamin K(GO:0032571) bone regeneration(GO:1990523) |
| 0.1 | 0.2 | GO:2000866 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.1 | 1.7 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.1 | 0.3 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.1 | 1.2 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.2 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) |
| 0.1 | 0.2 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.1 | 0.3 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 0.4 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.1 | 0.2 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 0.6 | GO:0097267 | omega-hydroxylase P450 pathway(GO:0097267) |
| 0.1 | 0.2 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.1 | 0.4 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.1 | 0.2 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.1 | 0.2 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.0 | 0.1 | GO:0006844 | acyl carnitine transport(GO:0006844) acyl carnitine transmembrane transport(GO:1902616) |
| 0.0 | 1.0 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.0 | 0.6 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.2 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.0 | 0.3 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.3 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.2 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 0.2 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.2 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.0 | 0.1 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.0 | 0.2 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.0 | 0.2 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.0 | 0.1 | GO:0071030 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.0 | 0.1 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.0 | 0.4 | GO:2000111 | senescence-associated heterochromatin focus assembly(GO:0035986) positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.0 | 0.1 | GO:0036512 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.0 | 0.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.6 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.3 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.4 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 1.0 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.4 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.0 | 0.2 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.8 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 | 0.3 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 4.1 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.1 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.0 | 0.2 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) |
| 0.0 | 0.2 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.1 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.0 | 0.1 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) |
| 0.0 | 1.1 | GO:0050912 | detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.0 | 0.1 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.1 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.0 | 0.2 | GO:0002371 | dendritic cell cytokine production(GO:0002371) |
| 0.0 | 0.4 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.4 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.1 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.3 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.4 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.3 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.1 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.2 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.0 | 0.2 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.0 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.3 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.9 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.2 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 1.5 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 0.3 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.2 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.1 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.1 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.0 | 0.1 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0061740 | protein targeting to lysosome involved in chaperone-mediated autophagy(GO:0061740) |
| 0.0 | 0.2 | GO:0070508 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.0 | 0.0 | GO:1990927 | negative regulation of synaptic vesicle recycling(GO:1903422) negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.0 | 0.8 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.2 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.0 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.1 | GO:0051272 | positive regulation of cell migration(GO:0030335) positive regulation of locomotion(GO:0040017) positive regulation of cellular component movement(GO:0051272) positive regulation of cell motility(GO:2000147) |
| 0.0 | 0.1 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.0 | 0.5 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.0 | GO:1902161 | transepithelial water transport(GO:0035377) positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.0 | 0.5 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.4 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.0 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.0 | 0.1 | GO:0003374 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.0 | 0.1 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.3 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.1 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.3 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.0 | GO:1903936 | response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.0 | 0.6 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.1 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.2 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.0 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.0 | 0.1 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.2 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.0 | GO:1902159 | regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 0.0 | 0.1 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.3 | 1.0 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.2 | 3.4 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.2 | 0.4 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.1 | 0.5 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 0.6 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 0.3 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.1 | 1.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 0.6 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.3 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.0 | 0.2 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.4 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.5 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.1 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.0 | 0.1 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.0 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.7 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.1 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.0 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.0 | 0.6 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.1 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.0 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.2 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 0.3 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.0 | GO:1990742 | microvesicle(GO:1990742) |
| 0.0 | 0.7 | GO:0001533 | cornified envelope(GO:0001533) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.4 | 1.1 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.3 | 1.0 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.2 | 1.0 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.2 | 0.7 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.1 | 6.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 1.0 | GO:0050294 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.1 | 0.5 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.6 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.1 | 0.3 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.1 | 0.8 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 1.0 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 3.1 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 0.5 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 0.3 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 0.3 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.1 | 0.4 | GO:0004040 | amidase activity(GO:0004040) |
| 0.1 | 0.2 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.1 | 0.3 | GO:1904493 | Ac-Asp-Glu binding(GO:1904492) tetrahydrofolyl-poly(glutamate) polymer binding(GO:1904493) |
| 0.1 | 1.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 1.8 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.8 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.4 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 0.2 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.1 | 0.2 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.9 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 1.0 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.1 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.0 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.2 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.3 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.2 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.0 | 0.2 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.9 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.1 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 3.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.2 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 1.0 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 1.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.3 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.1 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.0 | 0.2 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.2 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.4 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.4 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.2 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.1 | GO:0030760 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.4 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.2 | GO:0004407 | histone deacetylase activity(GO:0004407) protein deacetylase activity(GO:0033558) |
| 0.0 | 0.4 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.3 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0070283 | lipoate synthase activity(GO:0016992) radical SAM enzyme activity(GO:0070283) |
| 0.0 | 0.2 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.3 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.3 | GO:0030884 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 0.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.2 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.6 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.3 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.4 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0008513 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.1 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.0 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.0 | 0.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.1 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.2 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.1 | GO:0004075 | biotin carboxylase activity(GO:0004075) biotin binding(GO:0009374) |
| 0.0 | 0.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.2 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.0 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.0 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 2.0 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.0 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.0 | 0.2 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.0 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.4 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.3 | ST_IL_13_PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.4 | PID_IL5_PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.5 | PID_CONE_PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.4 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 3.5 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.8 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.0 | ST_T_CELL_SIGNAL_TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.6 | PID_NEPHRIN_NEPH1_PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.1 | ST_INTERFERON_GAMMA_PATHWAY | Interferon gamma pathway. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.1 | REACTOME_GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 3.4 | REACTOME_CS_DS_DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.8 | REACTOME_ROLE_OF_DCC_IN_REGULATING_APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 1.4 | REACTOME_TERMINATION_OF_O_GLYCAN_BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 1.1 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_GOLGI_MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 1.9 | REACTOME_METABOLISM_OF_STEROID_HORMONES_AND_VITAMINS_A_AND_D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.8 | REACTOME_IL_7_SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.6 | REACTOME_XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 1.0 | REACTOME_CYTOSOLIC_SULFONATION_OF_SMALL_MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.9 | REACTOME_HORMONE_SENSITIVE_LIPASE_HSL_MEDIATED_TRIACYLGLYCEROL_HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.6 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.6 | REACTOME_TRANSLOCATION_OF_ZAP_70_TO_IMMUNOLOGICAL_SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.5 | REACTOME_P2Y_RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 1.4 | REACTOME_PHASE_II_CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.3 | REACTOME_IL_6_SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.3 | REACTOME_RECYCLING_OF_BILE_ACIDS_AND_SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.0 | REACTOME_BINDING_AND_ENTRY_OF_HIV_VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.1 | REACTOME_NUCLEOTIDE_LIKE_PURINERGIC_RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.4 | REACTOME_IL_RECEPTOR_SHC_SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.4 | REACTOME_BASIGIN_INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.4 | REACTOME_GENERATION_OF_SECOND_MESSENGER_MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.2 | REACTOME_HDL_MEDIATED_LIPID_TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.5 | REACTOME_SEMA4D_INDUCED_CELL_MIGRATION_AND_GROWTH_CONE_COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.2 | REACTOME_INTRINSIC_PATHWAY | Genes involved in Intrinsic Pathway |